Self-organized multicellular structures from simple cell signaling: a computational model
Abstract
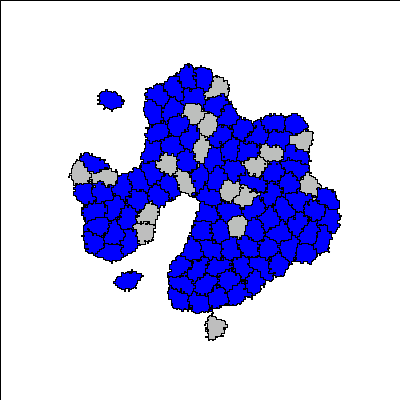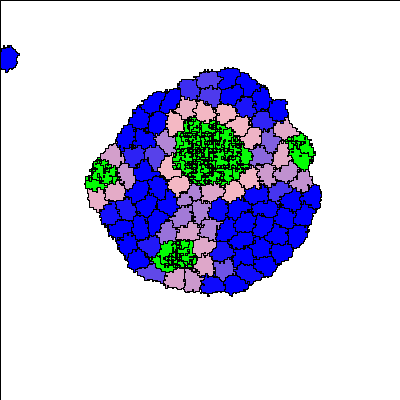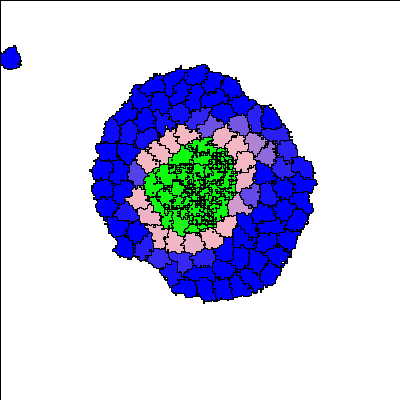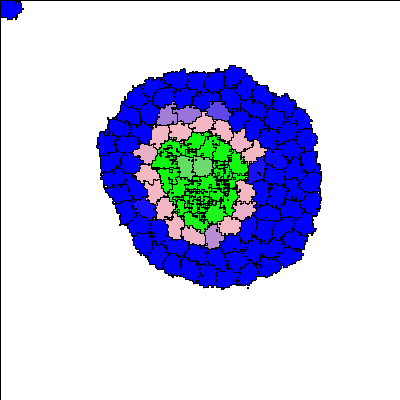
Recent synthetic biology experiments reveal that signaling modules designed to target cell-cell adhesion enable self-organization of multicellular structures [Toda, 2018]. Changes in homotypic adhesion that arise through contact-dependent signaling networks result in sorting of an aggregate into two- or three-layered structures. Here we investigate the formation, maintenance, and robustness of such self-organization in the context of a computational model. To do so, we use an established model for Notch/Ligand signaling within cells to set up differential E-cadherin expression. This signaling model is integrated with the Cellular Potts Model to track state changes, adhesion, and cell sorting in a group of cells. The resulting multicellular structures are in accordance with those observed in the experimental reference [Toda 2018]. In addition to reproducing these experimental results, we track the dynamics of the evolving structures and cell states to understand how such morphologies are dynamically maintained. This appears to be an important developmental principle that was not emphasized in previous models. Our computational model facilitates more detailed understanding of the link between intra- and intercellular signaling, spatio-temporal rearrangement, and emergent behaviour at the scale of hundred(s) of cells.