Poisson Disc Sampling
Poisson disc sampling is a technique to create blue noise spatial point patterns, i.e. patterns where all points are at least distance $r$ apart for some user-supplied density parameter Bridson 2007.
The new InitPoissonDisc initializer plugin allows you to initialize a tightly-packed population of cells based on this technique. This can be helpful to create initial conditions that closely resemble the properties of epithelial tissues.
InitPoissonDisc plugin
In Morpheus' implementation, you do not directly specify the density parameter or minimal distance. Instead, you configure the plugin by setting the total number of cells:
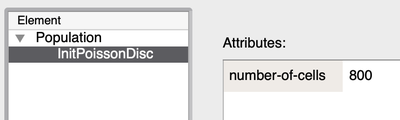
This is equivalent to this XML specification:
<CellPopulations>
<Population size="1" type="cells">
<InitPoissonDisc number-of-cells="800"/>
</Population>
</CellPopulations>
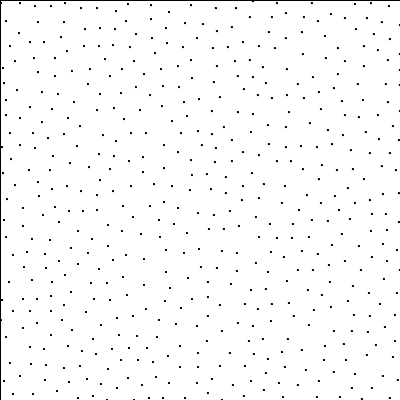
For a square lattice of size 400x400, this will create a point pattern like this:
Combining with InitVoronoi
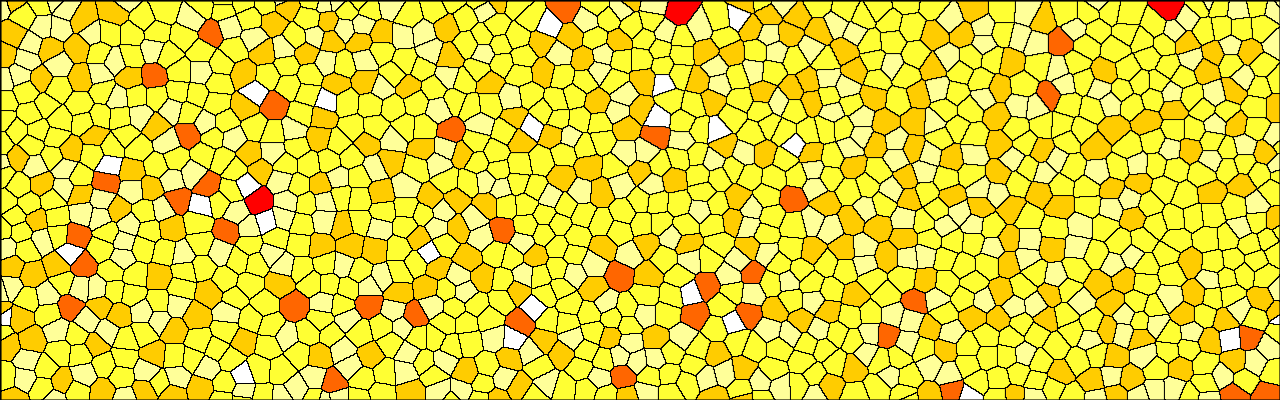
You can combine the InitPoissonDisc with the InitVoronoi plugin, which creates a Voronoi tesselation using the existing points as seeds. Note that the InitVoronoi must be specified after InitPoissonDisc to have effect.
<CellPopulations>
<Population size="1" type="cells">
<InitPoissonDisc number-of-cells="800"/>
<InitVoronoi/>
</Population>
</CellPopulations>
With this combination, you can create the following initial cellular configuration:
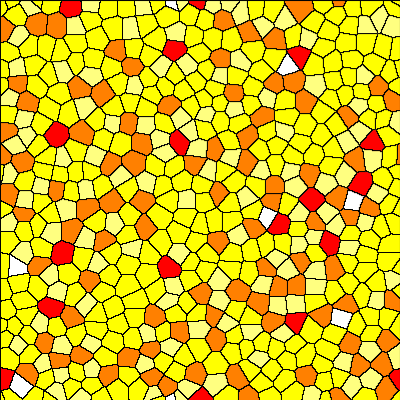
Interestingly, this produces tesselations that have properties closely resembling epithelial tissues as seen in the following histograms of the area per cell (cell.volume) and number of neighbors:
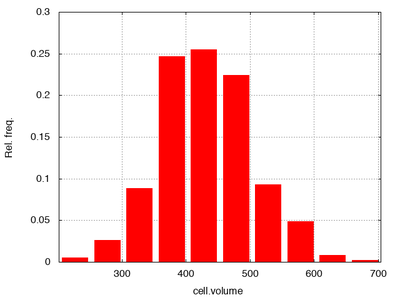
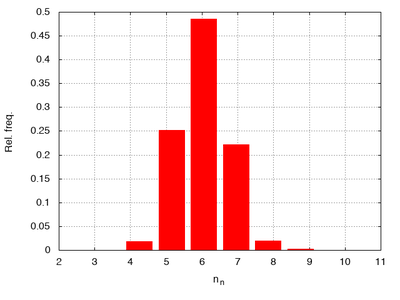
Counting neighbors using local.cell.id
As an useful aside: Counting the number of neighboring cells is done here using NeighborhoodReporter that reports, for each cell, the sum over all neighboring cells for which the cell.id is not equal to its own cell id local.cell.id:
<CellType class="biological" name="cells">
<Property symbol="n_n" value="0.0" name="Number of neighbors"/>
<NeighborhoodReporter>
<Input scaling="cell" value="local.cell.id != cell.id "/>
<Output symbol-ref="n_n" mapping="sum"/>
</NeighborhoodReporter>
</CellType>
The concept of comparing to the value of a cells own property (local.cell.id) is also new in Morpheus 2.1.
Conclusion
The new InitPoissonDisc creates point patterns with a bounded minimal distance between cell centers and can be nicely combined with InitVoronoi to generate realistic epithelium-like tesselations that can serve as initial conditions for your models.