MembraneProperties: Cell Polarization and Chemotaxis
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Introduction
This model of cell polarity shows the coupling of three model formalisms:
- A cellular Potts model with
DirectedMotionthat responds to cell polarity (but not directly to the external signal gradient), - a co-moving PDE model on the membrane of the cell (to let cell polarity emerge from distributed receptor-signal interactions) and
- an external signal gradient (implemented as a global field with random fluctuations).
The cell membrane polarizes in response to the external signal U that is sensed as a pattern (of receptor activation) s along the cell membrane.
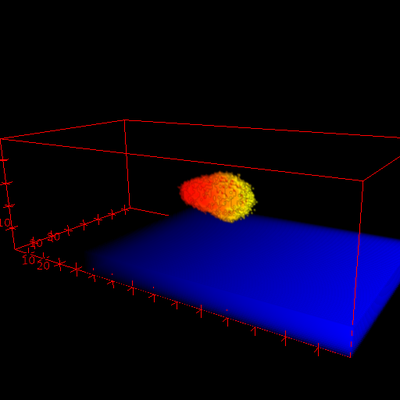
Description
This example implements two models of cell polarity formation: Meinhardt’s substrate-depletion model (ASDM) and Edelstein-Keshet’s wave-pinning (WP) model. The user can switch the polarity model by disabling/enabling the relevant System under CellType. Equations and parameter values are copied from Mori et al..
Each model defines a two-component reaction-diffusion system (of MembraneProperty A and B) representing membrane-bound molecules, and is mapped to and co-moves with a cell. Cell motility and cell shape dynamics are given by the cellular Potts model. An external gradient with random fluctuations, specified in Global, provides a signal for the polarization of the cell. In turn, the pattern of the MembraneProperty A gives the polarity vector of the cell. The polarity vector then controls the DirectedMotion behavior of the cell. A TiffPlotter exports the full 3D plus time data set.
After a switch in direction of the gradient, the cell re-polarizes in the new direction and starts to move in the new direction, if the wave pinning model has been selected (see video below). A GnuPlotter visualizes the gradient, cell shape and MembraneProperty A at a horizontal slice through 3D space (shown below) and a Logger visualizes the (phi, theta)-dependency of the MembraneProperty A (not shown).
The alternative ASDM maintains its previous polarity for much longer and does not re-polarize during the runtime (see video below). On a longer time scale, the ASDM may eventually re-polarize the 3D cell by rotating the activator maximum over the cell membrane. The cell would take a U-turn with a large radius. The same behaviours are observed for any other strength of the gradient’s fluctuations as well as for a smooth gradient.
Reference
This model reproduces a published result, originally obtained with a different simulator:
Y. Mori, A. Jilkine, L. Edelstein-Keshet: Wave-Pinning and Cell Polarity from a Bistable Reaction-Diffusion System.. Biophysical Journal 94(9): 3684-3697, 2008.
Model
Examples → Multiscale → CellPolarity.xml or
CellPolarity.xml
XML Preview
<MorpheusModel version="4">
<Description>
<Title>Example-CellPolarity</Title>
<Details>Chemotaxis of polarized cell
----------------------------
Show feedback between cell motility, cell polarization and external gradient.
Two cell polarization models are taken from the original publication:
Y. Mori, A. Jilkine and L. Edelstein-Keshet (2008). Wave-Pinning and Cell Polarity from a Bistable Reaction-Diffusion System. Biophysical Journal 94(9), 3684-3697. DOI:10.1529/biophysj.107.120824
and can be enabled/disabled for a cell in 3D through the respective System in the Celltype 'cells':
- Substrate-depletion model: no repolarization
- Wave-pinning model: repolarization (as studied in detail in the above publication)
Events in Global:
- Switch on/off and change gradients (noise through global constant)</Details>
</Description>
<Global>
<Field symbol="U" value="noisy(l.x / lattice.x)">
<Diffusion rate="0.0"/>
</Field>
<Event trigger="on-change" time-step="100">
<Condition>time>=1000</Condition>
<Rule symbol-ref="U">
<Expression>noisy(0.5)</Expression>
</Rule>
</Event>
<Event trigger="on-change" time-step="100">
<Condition>time>=1500</Condition>
<Rule symbol-ref="U">
<Expression>noisy((lattice.x-l.x) / lattice.x)</Expression>
</Rule>
</Event>
<Constant symbol="l_U" value="0.0"/>
<Constant symbol="lengthscale" value="3"/>
<Constant symbol="noise" value="0.2"/>
<Function symbol="noisy">
<Parameter symbol="v" name="value"/>
<Expression>max(0,(v+rand_norm(0,noise)))</Expression>
</Function>
</Global>
<Space>
<Lattice class="cubic">
<Size symbol="lattice" value="150, 75, 30"/>
<BoundaryConditions>
<Condition type="noflux" boundary="x"/>
<Condition type="periodic" boundary="y"/>
<Condition type="constant" boundary="z"/>
</BoundaryConditions>
<NodeLength value="1.0"/>
<Neighborhood>
<Distance>2.0</Distance>
</Neighborhood>
</Lattice>
<SpaceSymbol symbol="l"/>
<MembraneLattice>
<Resolution symbol="memsize" value="50"/>
<SpaceSymbol symbol="m"/>
</MembraneLattice>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="2500"/>
<SaveInterval value="0"/>
<RandomSeed value="0"/>
<TimeSymbol symbol="time"/>
</Time>
<CellTypes>
<CellType name="cells" class="biological">
<VolumeConstraint target="5000" strength="0.1"/>
<SurfaceConstraint target="1" strength="0.02" mode="aspherity"/>
<MembraneProperty symbol="A" name="A" value="1.5 + rand_uni(-0.1,0.1)">
<Diffusion rate="0.05*lengthscale^2"/>
</MembraneProperty>
<MembraneProperty symbol="B" name="B" value="0.8">
<Diffusion rate="10*lengthscale^2"/>
</MembraneProperty>
<MembraneProperty symbol="c" name="chemotactic strength" value="0">
<Diffusion rate="0"/>
</MembraneProperty>
<MembraneProperty symbol="s" name="signal" value="0">
<Diffusion rate="0"/>
</MembraneProperty>
<System time-step="0.05" name="WavePinning" solver="Dormand-Prince [adaptive, O(5)]">
<Constant symbol="k_0" value="0.067"/>
<Constant symbol="gamma" value="1"/>
<Constant symbol="delta" value="1"/>
<Constant symbol="K" value="1"/>
<!-- <Disabled>
<Constant symbol="sigma" name="spatial-length-signal" value="5.0"/>
</Disabled>
-->
<Constant symbol="n" name="Hill coefficient" value="2"/>
<Intermediate symbol="F" value="B*(k_0+ 0.2*(s-U.mean) + (gamma*A^n) / (K^n + A^n) ) - delta*A"/>
<DiffEqn symbol-ref="A">
<Expression>F</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B">
<Expression>-F</Expression>
</DiffEqn>
<Rule symbol-ref="c">
<Expression>A^2*1000</Expression>
</Rule>
</System>
<Mapper name="Sensing the field">
<Input value="U"/>
<Output symbol-ref="s" mapping="average"/>
</Mapper>
<PropertyVector symbol="p" name="polarity" value="0.0, 0.0, 0.0"/>
<Mapper name="Polarity extraction from membrane">
<Input value="A"/>
<Polarity symbol-ref="p"/>
</Mapper>
<DirectedMotion strength="0.04" direction="p"/>
<!-- <Disabled>
<System time-step="0.05" name="Substrate-Depletion(ASDM)" solver="Dormand-Prince [adaptive, O(5)]">
<Rule symbol-ref="c">
<Expression>A^2*5e1</Expression>
</Rule>
<DiffEqn symbol-ref="A">
<Expression>B*(s.norm*A^2/(1+s_a*A^2)) - r_a * A</Expression>
</DiffEqn>
<DiffEqn symbol-ref="B">
<Expression>b_b - B*(s.norm*A^2/(1+s_a*A^2)) - r_b*B</Expression>
</DiffEqn>
<Constant symbol="s_a" value="0.1"/>
<Constant symbol="r_a" value="0.1"/>
<Constant symbol="b_b" value="0.15"/>
<Constant symbol="r_b" value="0.0"/>
<Function symbol="s.norm">
<Expression>0.1+0.01*(s-U.mean)/U.mean</Expression>
</Function>
<Disabled>
<Constant symbol="s.norm" value="0.1"/>
</Disabled>
</System>
</Disabled>
-->
<Property symbol="U.mean" value="0.0"/>
<Mapper name="scalar signal average over whole cell">
<Input value="U"/>
<Output symbol-ref="U.mean" mapping="average"/>
</Mapper>
</CellType>
</CellTypes>
<CPM>
<Interaction default="0.0"/>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<MetropolisKinetics yield="0.05" temperature="1"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>6</Order>
</Neighborhood>
</ShapeSurface>
</CPM>
<CellPopulations>
<Population type="cells" size="0">
<InitCellObjects mode="distance">
<Arrangement repetitions="1, 1, 1" displacements="1, 1, 1">
<Sphere center="25 37 15" radius="10"/>
</Arrangement>
</InitCellObjects>
</Population>
</CellPopulations>
<Analysis>
<Gnuplotter time-step="50">
<Terminal name="png"/>
<Plot slice="15">
<Field symbol-ref="U"/>
<Cells min="0.2" value="A"/>
<CellArrows orientation="p * 50"/>
</Plot>
</Gnuplotter>
<TiffPlotter time-step="100" format="8bit" OME-header="false">
<Channel symbol-ref="cell.id" celltype="cells"/>
<Channel symbol-ref="c" celltype="cells"/>
<Channel symbol-ref="U"/>
</TiffPlotter>
<Logger time-step="50">
<Input>
<Symbol symbol-ref="A"/>
</Input>
<Output>
<TextOutput file-format="matrix"/>
</Output>
<Plots>
<SurfacePlot time-step="50">
<Color-bar>
<Symbol symbol-ref="A"/>
</Color-bar>
<Terminal terminal="png"/>
</SurfacePlot>
</Plots>
<Restriction>
<Celltype celltype="cells"/>
</Restriction>
</Logger>
<ModelGraph include-tags="#untagged" format="svg" reduced="false"/>
</Analysis>
</MorpheusModel>

Downloads
Files associated with this model: