Hepatocellular Carcinoma
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Hepatocyte–sinusoid alignment (HSA) causes asymmetric shapes in early hepatocellular tumors.
Introduction
The mammalian liver is subdivided into lobes and further into thousands of hexagonal columns, called lobules, which act in parallel. Blood enters a lobule at the periphery, flows through a network of sinusoids and leaves at the center. The sinusoids may also prepattern the growth of cell clones. This model studies the influence of hepatocyte–sinusoid alignment (HSA), i.e. the orientation of the division plane such that daughter hepatocytes align with nearby sinusoids, on the shape of tumor nodules in liver lobules (Fig. 1).
: [**Höhme _et al._**](#reference), [Fig. 5](https://link.springer.com/article/10.1007/s11538-017-0375-1/figures/5)). The data show a decreasing aspect ratio of long over short cluster axes from early to late stages.](/media/model/m9148/hoehme-2018_fig-5_rat-liver_hu512b0ef0b563c93d5d610ae4bebe448a_731249_43bad210eed7c45384dcb0d2448e2310.jpg)
Description
As the original model for early hepatocellular carcinoma (Fig. 2) had been constructed by extending a model for liver regeneration by Höhme et al., 2010, we here equally extended the corresponding MorpheusML model (M9147).
While the original model considered a three-dimensional liver lobule, the MorpheusML model currently (only) represents the two-dimensional cross section with correspondingly lower cell numbers. Also the total simulation duration is reduced to 7 days compared to 10 days in the original model. Starting with only one tumor cell in the liver lobule embedded in 675 hepatocytes (instead of around 4000 hepatocytes in the original model), the tumor gains 400 tumor cells by the end of the simulated time.
All other parameters and observables are comparable. At time $0\ \mathrm{d}$ (after the initialization period finished), a single hepatocyte switches to a high proliferation mode and its daughter cells inherit this parameter set, forming a tumor clone. To compare tumor shapes within a single simulation run, two tumors are initialized, one placed in the lower left with HSA and another in the top right without HSA. The aspect ratio of long and short shape axes is approximately measured and visualized by the model itself.
: [**Höhme _et al._**](#reference), [Fig. 3](https://link.springer.com/article/10.1007/s11538-017-0375-1/figures/3))](/media/model/m9148/hoehme-2018_fig-3_tumor-growth-scenarios_hu167fa88d048570624f536ddd4b34fbf3_2396240_4a0cf322fd7986a4c24b4ba679b447e7.jpg)
Results
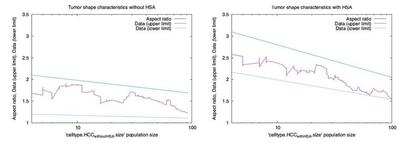
The Morpheus model qualitatively reproduces the original model (see Video 1). HSA (light purple cells) causes early asymmetry of tumor cell clusters that decreases later. The time course of the aspect ratio was quantified in Höhme et al. and is shown in Fig. 3. Likewise, this time course has been quantified for the Morpheus simulations and plotted in Fig. 4 (noisy curves from a single realization) and compared to the upper and lower limits from the original study (straight lines).
 Comparison of the ratio of the longest versus shortest tumor axis. Black: With HSA. Red: No HSA. ([CC BY 4.0](https://creativecommons.org/licenses/by/4.0/): [**Höhme _et al._**](#reference), [Fig. 4](https://link.springer.com/article/10.1007/s11538-017-0375-1/figures/4))](/media/model/m9148/hoehme-2018_fig-4_tumor-axis-ratios_hu2f55a1a46bdc9a39fc2c3d8363e96e42_228643_e021068b2864ad30dddaef9d0ffb98d6.jpg)
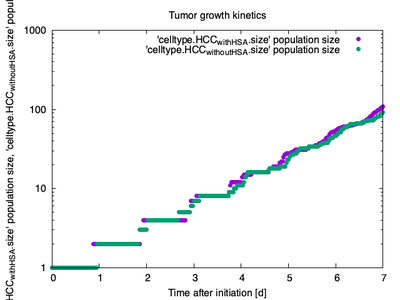
Due to the increased competition for space in the two-dimensional cross section compared to the three-dimensional space in the published model, tumors grow slightly slower in the reproduced model (Fig. 6) than in the original model (Fig. 5). Nonetheless, the exponential form of the growth kinetics is reproduced as well as the slightly faster growth of clones with HSA.
 Growth kinetics of tumors in case of HSA (black) and no HSA (red) ([CC BY 4.0](https://creativecommons.org/licenses/by/4.0/): [**Höhme _et al._**](#reference), [Fig. 14](https://link.springer.com/article/10.1007/s11538-017-0375-1/figures/14))](/media/model/m9148/hoehme-2018_fig-14_tumor-growth-kinetics_hu558fe4e7d67df0602cb26165b6508d6e_49064_b49ac881a6d6f3affde14bb71cd5e228.jpg)
Reference
This model reproduces a published result, originally obtained with a different simulator:
S. Höhme, F. Bertaux, W. Weens, B. Grasl-Kraupp, J. G. Hengstler, D. Drasdo: Model Prediction and Validation of an Order Mechanism Controlling the Spatiotemporal Phenotype of Early Hepatocellular Carcinoma. Bull. Math. Biol. 80: 1134–1171, 2018.
Model
M9148_hcc_model.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="4">
<Time>
<StartTime value="0"/>
<StopTime value="2160">
<Annotation>[time] = 5 min, i. e. 288 time units correspond to 24 h; total time (StopTime) of 2160 units gives 0.5 d burn in + 7 d dynamics (to gain 400 cells in clone)</Annotation>
</StopTime>
<TimeSymbol symbol="time" name="time"/>
<RandomSeed value="0"/>
</Time>
<Description>
<Title>M9148 HCC</Title>
<Details>Model ID: https://identifiers.org/morpheus/M9148
Title: Hepatocellular Carcinoma
Authors: S. Höhme, F. Bertaux, W. Weens, B. Grasl-Kraupp, J. G. Hengstler, D. Drasdo
Submitters: L. Brusch, D. Jahn
Curators: L. Brusch, D. Jahn
Date: 12.04.2024
Software: Morpheus (open source). Download from: https://morpheus.gitlab.io
Units: [time] = 1 d
[space] = 3 µm
Reference: This model reproduces a published result, originally obtained with a different simulator:
S. Höhme, F. Bertaux, W. Weens, B. Grasl-Kraupp, J. G. Hengstler, D. Drasdo: Model Prediction and Validation of an Order Mechanism Controlling the Spatiotemporal Phenotype of Early Hepatocellular Carcinoma. Bull. Math. Biol. 80, 1134–1171, 2018.
https://doi.org/10.1007/s11538-017-0375-1
</Details>
</Description>
<Space>
<SpaceSymbol symbol="l" name="location"/>
<Lattice class="hexagonal">
<Size symbol="size" value="200, 200, 0"/>
<BoundaryConditions>
<Condition type="noflux" boundary="x"/>
<Condition type="noflux" boundary="y"/>
</BoundaryConditions>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<Annotation>Lobule radius in 2D slice (lit.): 284.3 µm
Hexagon diameter: 284.3 µm * 2 / node.length = 190</Annotation>
<Domain boundary-type="noflux">
<Hexagon diameter="190"/>
</Domain>
<NodeLength symbol="um" value="3"/>
</Lattice>
<MembraneLattice>
<Resolution value="100"/>
<SpaceSymbol symbol="membrane"/>
</MembraneLattice>
</Space>
<Global>
<Constant symbol="apical.polarization" tags="CCl4, HCC" value="0">
<Annotation>0: tangential
1: radial</Annotation>
</Constant>
<Constant symbol="node.length" name="Lattice node length in µm" value="3"/>
<Constant symbol="node.volume" value="node.length^2 * sqrt(3)/2 * 10^-6"/>
<Constant symbol="time_span.day" value="12 * 24">
<Annotation>Factor to scale time from 5 min steps to 1 d</Annotation>
</Constant>
<Function symbol="time.days" name="Time in days">
<Expression>time/time_span.day</Expression>
</Function>
<Constant symbol="time.burn_in" tags="HCC" value="0.50">
<Annotation>Initialization time in days</Annotation>
</Constant>
<Function symbol="time.post_init" name="Time after initiation [d]" tags="HCC">
<Annotation>Time in days excluding initialization time (0 ≤ t ≤ time.cell_death). Used for plotting.</Annotation>
<Expression>time.days - time.burn_in</Expression>
</Function>
<ConstantVector symbol="size.physical" tags="CCl4, HCC" value="size.x, size.y * sqrt(3)/2, size.z"/>
<VectorField symbol="sinusoid" tags="CCl4, HCC" value="l - size.physical/2"/>
<ConstantVector symbol="hepatocyte.sinusoid_mean" tags="CCl4, HCC" value="0.0, 0.0, 0.0"/>
<Constant symbol="membrane.apical" tags="CCl4, HCC" value="0.0">
<Annotation>Default value for apical adhesive. Value is set per CellType.</Annotation>
</Constant>
<Constant symbol="hepatocyte.contactEnergy" value="-0.5">
<Annotation>Common value for all contact energies with hepatocytes involved</Annotation>
</Constant>
<Constant symbol="hepatocyte.adhesionStrength" value="4.0">
<Annotation>Common value for //CPM/Interaction/Contact/HomophilicAdhesion/@strength of all possible hepatocyte interactions</Annotation>
</Constant>
<Constant symbol="hepatocyte.time_span.cell_cycle" name="tau" tags="CCl4, HCC" value="1">
<Annotation>Time of one cell cycle in days (Table 2 in Hoehme et al. 2018)</Annotation>
</Constant>
<Field symbol="c" name="Morphogen concentration" tags="CCl4, HCC" value="0">
<Annotation>Diffusion constant converted from cm^2/s to 3 µm^2/5 min:
10e-6 cm^2/s = 10e-6 * (10^4)^2 µm^2/s = 10e-6 * (10^4)^2 / 3^2 (3 µm)^2/s = 10^-6 * 10^8 / 9 * 300 (3 µm)^2/5 min = 3333.3</Annotation>
<Diffusion rate="33"/>
</Field>
<Field symbol="hepatocyte.lesion.pattern" name="Hepatocyte lesion pattern" tags="CCl4, HCC" value="false"/>
<Field symbol="hepatocyte.lesion.pattern_including_cv" name="Hepatocyte lesion pattern plus CV cross section" tags="CCl4, HCC" value="false"/>
<Constant symbol="c.decay" tags="CCl4" value="0">
<Annotation>Global initialization of decay rate. Actual CCl4 depletion occurs only in CellType 'hepatocytes'.</Annotation>
</Constant>
<System time-step="mcs_duration" name="CCl4 input and induced morphogen formation, diffusion and depletion" solver="Bogacki-Shampine [adaptive, O(3)]" tags="CCl4, HCC">
<Rule symbol-ref="hepatocyte.lesion.pattern" tags="CCl4, HCC">
<Expression>cell.type == celltype.medium.id</Expression>
</Rule>
<Rule symbol-ref="hepatocyte.lesion.pattern_including_cv" tags="CCl4, HCC">
<Expression>cell.type == celltype.medium.id or cell.type == celltype.CV.id</Expression>
</Rule>
<Constant symbol="c.target" tags="CCl4, HCC" value="1"/>
<DiffEqn symbol-ref="c" tags="CCl4, HCC">
<Expression>-c.decay*c + (c.target-c)*hepatocyte.lesion.pattern_including_cv</Expression>
</DiffEqn>
</System>
<Constant symbol="clone.celldiameter" name="23.3+-3.1 µm from Table 1 in Hoehme et al. 2018" tags="HCC, CCl4" value="23.3 / node.length"/>
<Variable symbol="clone.withHSA.area" tags="HCC" value="0.0"/>
<Variable symbol="clone.withHSA.inner" tags="HCC" value="0.0"/>
<Variable symbol="clone.withHSA.outer" tags="HCC" value="0.0"/>
<Mapper time-step="10" name="clone.withHSA.area" tags="HCC">
<Input value="node.volume*cell.volume*(cell.type==celltype.HCC_with_HSA.id)"/>
<Output symbol-ref="clone.withHSA.area" mapping="sum"/>
</Mapper>
<Mapper time-step="10" name="clone.withHSA.inner" tags="HCC">
<Input value="if(cell.type==celltype.HCC_with_HSA.id,hepatocyte.sinusoid_mean.abs,size.x+size.y)"/>
<Output symbol-ref="clone.withHSA.inner" mapping="minimum"/>
</Mapper>
<Mapper time-step="10" name="clone.withHSA.outer" tags="HCC">
<Input value="if(cell.type==celltype.HCC_with_HSA.id,hepatocyte.sinusoid_mean.abs,0)"/>
<Output symbol-ref="clone.withHSA.outer" mapping="maximum"/>
</Mapper>
<Function symbol="clone.withHSA.radialspan" tags="HCC">
<Expression>node.length*10^-3*(clone.withHSA.outer-clone.withHSA.inner+clone.celldiameter)/2</Expression>
</Function>
<Function symbol="clone.withHSA.tangentialspan" tags="HCC">
<Expression>clone.withHSA.area/(pi*clone.withHSA.radialspan)</Expression>
</Function>
<Function symbol="clone.withHSA.aspectratio" name="Aspect ratio" tags="HCC">
<Expression>max(clone.withHSA.radialspan,clone.withHSA.tangentialspan)/min(clone.withHSA.radialspan,clone.withHSA.tangentialspan)</Expression>
</Function>
<Variable symbol="clone.withoutHSA.area" tags="HCC" value="0.0"/>
<Variable symbol="clone.withoutHSA.inner" tags="HCC" value="0.0"/>
<Variable symbol="clone.withoutHSA.outer" tags="HCC" value="0.0"/>
<Mapper time-step="10" name="clone.withoutHSA.area" tags="HCC">
<Input value="node.volume*cell.volume*(cell.type==celltype.HCC_without_HSA.id)"/>
<Output symbol-ref="clone.withoutHSA.area" mapping="sum"/>
</Mapper>
<Mapper time-step="10" name="clone.withoutHSA.inner" tags="HCC">
<Input value="if(cell.type==celltype.HCC_without_HSA.id,hepatocyte.sinusoid_mean.abs,size.x+size.y)"/>
<Output symbol-ref="clone.withoutHSA.inner" mapping="minimum"/>
</Mapper>
<Mapper time-step="10" name="clone.withoutHSA.outer" tags="HCC">
<Input value="if(cell.type==celltype.HCC_without_HSA.id,hepatocyte.sinusoid_mean.abs,0)"/>
<Output symbol-ref="clone.withoutHSA.outer" mapping="maximum"/>
</Mapper>
<Function symbol="clone.withoutHSA.radialspan" tags="HCC">
<Expression>node.length*10^-3*(clone.withoutHSA.outer-clone.withoutHSA.inner+clone.celldiameter)/2</Expression>
</Function>
<Function symbol="clone.withoutHSA.tangentialspan" tags="HCC">
<Expression>clone.withoutHSA.area/(pi*clone.withoutHSA.radialspan)</Expression>
</Function>
<Function symbol="clone.withoutHSA.aspectratio" name="Aspect ratio" tags="HCC">
<Expression>max(clone.withoutHSA.radialspan,clone.withoutHSA.tangentialspan)/min(clone.withoutHSA.radialspan,clone.withoutHSA.tangentialspan)</Expression>
</Function>
<Constant symbol="hepatocyte.time_span.cell_cycle_delta" tags="HCC" value="4 / 24">
<Annotation>Delta tau [d] = 2h from Ad(A-5) in Hoehme et al. 2018, but *2 to match Fig.14 inset</Annotation>
</Constant>
</Global>
<CellTypes>
<CellType class="medium" name="medium" tags="CCl4, HCC"/>
<CellType class="biological" name="CV" tags="CCl4, HCC">
<FreezeMotion tags="CCl4, HCC">
<Condition>1</Condition>
</FreezeMotion>
</CellType>
<CellType class="biological" name="PV" tags="CCl4, HCC">
<FreezeMotion tags="CCl4, HCC">
<Condition>1</Condition>
</FreezeMotion>
</CellType>
<CellType class="biological" name="hepatocytes" tags="CCl4, HCC">
<Property symbol="hepatocyte.time.birth" tags="CCl4, HCC" value="-hepatocyte.time_span.cell_cycle"/>
<VolumeConstraint target="pi * (0.5*23.3/node.length)^2 / (sqrt(3)/2)" name="Target area (2D)" strength="1" tags="CCl4, HCC">
<Annotation>Cell diameter (lit.): 23.3 µm
</Annotation>
</VolumeConstraint>
<SurfaceConstraint target="1" name="Target perimenter (2D)" strength="1" tags="CCl4, HCC" mode="aspherity"/>
<Constant symbol="c.decay" tags="CCl4" value="0.004">
<Annotation>CCl4 depletion occurs only in hepatocytes, i.e. CCl4 decay rate is 0 at in all other locations.</Annotation>
</Constant>
<PropertyVector symbol="hepatocyte.sinusoid_mean" tags="CCl4, HCC" value="0.0, 0.0, 0.0"/>
<VectorMapper tags="CCl4, HCC">
<Input value="sinusoid"/>
<Output symbol-ref="hepatocyte.sinusoid_mean" mapping="average"/>
</VectorMapper>
<MembraneProperty symbol="membrane.apical" tags="CCl4, HCC" value="0">
<Diffusion rate="0.0"/>
</MembraneProperty>
<Equation symbol-ref="membrane.apical" tags="CCl4, HCC">
<Annotation>Assign the value "1" to the MembraneProperty on apical sides (± 45°; ± π/4) of the hepatocytes. Needed for homophilic adhesion.
- Use cos() version for apical sides perpendicular to sinusoids (tangential).
- Use sin() version for apical sides parallel to sinusoids (radial).</Annotation>
<Expression>apical.polarization == 0 ? (abs(cos(membrane.phi - hepatocyte.sinusoid_mean.phi)) <= sin(pi/4) ? 1 : 0) : (abs(sin(membrane.phi - hepatocyte.sinusoid_mean.phi)) <= sin(pi/4) ? 1 : 0)</Expression>
</Equation>
<Chemotaxis strength="10" tags="CCl4, HCC" field="c"/>
</CellType>
<CellType class="biological" name="HCC_with_HSA" tags="HCC">
<Property symbol="hepatocyte.time.birth" tags="HCC" value="time.burn_in"/>
<VolumeConstraint target="pi * (0.5*23.3/node.length)^2 / (sqrt(3)/2)" name="Target area (2D)" strength="1" tags="CCl4, HCC">
<Annotation>Cell diameter (lit.): 23.3 µm
</Annotation>
</VolumeConstraint>
<SurfaceConstraint target="1" name="Target perimenter (2D)" strength="1" tags="CCl4, HCC" mode="aspherity"/>
<PropertyVector symbol="hepatocyte.sinusoid_mean" tags="CCl4, HCC" value="0.0, 0.0, 0.0"/>
<VectorMapper tags="CCl4, HCC">
<Input value="sinusoid"/>
<Output symbol-ref="hepatocyte.sinusoid_mean" mapping="average"/>
</VectorMapper>
<MembraneProperty symbol="membrane.apical" tags="CCl4, HCC" value="0">
<Diffusion rate="0.0"/>
</MembraneProperty>
<Equation symbol-ref="membrane.apical" tags="CCl4, HCC">
<Annotation>Assign the value "1" to the MembraneProperty on apical sides (± 45°; ± π/4) of the hepatocytes. Needed for homophilic adhesion.
- Use cos() version for apical sides perpendicular to sinusoids (tangential).
- Use sin() version for apical sides parallel to sinusoids (radial).</Annotation>
<Expression>apical.polarization == 0 ? (abs(cos(membrane.phi - hepatocyte.sinusoid_mean.phi)) <= sin(pi/4) ? 1 : 0) : (abs(sin(membrane.phi - hepatocyte.sinusoid_mean.phi)) <= sin(pi/4) ? 1 : 0)</Expression>
</Equation>
<!-- <Disabled>
<Constant symbol="hepatocyte.proliferation_p" name="division rate from Fig.14 in Hoehme et al. 2018: 1/(1080min/(5min/timestep))=1/(216 time steps)" tags="HCC" value="1/216"/>
</Disabled>
-->
<!-- <Disabled>
<System time-step="12" solver="Euler [fixed, O(1)]" tags="CCl4">
<Rule symbol-ref="hepatocyte.proliferation_p" tags="CCl4">
<Annotation>Definition of the cell division probability of BrdU-positive hepatocytes depending on the the layer number.</Annotation>
<Expression>if(time.regeneration >= 2.9 - hepatocyte.time_span.proliferation/2 && time.regeneration < 2.9 + hepatocyte.time_span.proliferation/2,
if(hepatocyte.layer.self == 1, 0.3,
if(hepatocyte.layer.self == 2, 0.25,
if(hepatocyte.layer.self == 3, 0.2,
if(hepatocyte.layer.self == 4, 0.2, 0.1)))), 0)</Expression>
</Rule>
</System>
</Disabled>
-->
<Property symbol="hepatocyte.time.division" tags="HCC" value="hepatocyte.time_span.cell_cycle"/>
<Property symbol="hepatocyte.time.nextdivision" tags="HCC" value="rand_norm(hepatocyte.time_span.cell_cycle,hepatocyte.time_span.cell_cycle_delta)"/>
<CellDivision trigger="on-change" orientation="hepatocyte.sinusoid_mean" division-plane="oriented" name="proliferation w/ HSA" tags="CCl4, HCC">
<Condition>time.days >= hepatocyte.time.birth+hepatocyte.time.division</Condition>
<Triggers>
<Rule symbol-ref="hepatocyte.time.birth">
<Expression>time.days</Expression>
</Rule>
<Rule symbol-ref="hepatocyte.time.nextdivision" tags="HCC">
<Expression>rand_norm(hepatocyte.time_span.cell_cycle,hepatocyte.time_span.cell_cycle_delta)</Expression>
</Rule>
</Triggers>
</CellDivision>
<!-- <Disabled>
<Event trigger="when-true" name="truncate Gaussian for individual cell cycle duration" tags="HCC">
<Condition>time.days < hepatocyte.time.birth+2*mcs_duration</Condition>
<Rule symbol-ref="hepatocyte.time.division" tags="HCC">
<Expression>if( hepatocyte.time.nextdivision < (hepatocyte.time_span.cell_cycle-hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle-hepatocyte.time_span.cell_cycle_delta), if( hepatocyte.time.nextdivision > (hepatocyte.time_span.cell_cycle+hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle+hepatocyte.time_span.cell_cycle_delta), hepatocyte.time.nextdivision))</Expression>
</Rule>
</Event>
</Disabled>
-->
<System time-step="mcs_duration" name="truncate Gaussian for individual cell cycle duration" solver="Euler [fixed, O(1)]" tags="HCC">
<Rule symbol-ref="hepatocyte.time.division" tags="HCC">
<Expression>if(hepatocyte.time.nextdivision < (hepatocyte.time_span.cell_cycle - hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle - hepatocyte.time_span.cell_cycle_delta),
if(hepatocyte.time.nextdivision > (hepatocyte.time_span.cell_cycle + hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle + hepatocyte.time_span.cell_cycle_delta), hepatocyte.time.nextdivision))</Expression>
</Rule>
</System>
<!-- <Disabled>
<Chemotaxis strength="100" tags="CCl4" field="c"/>
</Disabled>
-->
</CellType>
<CellType class="biological" name="HCC_without_HSA" tags="HCC">
<Property symbol="hepatocyte.time.birth" tags="HCC" value="time.burn_in"/>
<VolumeConstraint target="pi * (0.5*23.3/node.length)^2 / (sqrt(3)/2)" name="Target area (2D)" strength="1" tags="CCl4, HCC">
<Annotation>Cell diameter (lit.): 23.3 µm
</Annotation>
</VolumeConstraint>
<SurfaceConstraint target="1" name="Target perimenter (2D)" strength="1" tags="CCl4, HCC" mode="aspherity"/>
<PropertyVector symbol="hepatocyte.sinusoid_mean" tags="CCl4, HCC" value="0.0, 0.0, 0.0"/>
<VectorMapper tags="CCl4, HCC">
<Input value="sinusoid"/>
<Output symbol-ref="hepatocyte.sinusoid_mean" mapping="average"/>
</VectorMapper>
<MembraneProperty symbol="membrane.apical" tags="CCl4, HCC" value="0">
<Diffusion rate="0.0"/>
</MembraneProperty>
<Equation symbol-ref="membrane.apical" tags="CCl4, HCC">
<Annotation>Assign the value "1" to the MembraneProperty on apical sides (± 45°; ± π/4) of the hepatocytes. Needed for homophilic adhesion.
- Use cos() version for apical sides perpendicular to sinusoids (tangential).
- Use sin() version for apical sides parallel to sinusoids (radial).</Annotation>
<Expression>apical.polarization == 0 ? (abs(cos(membrane.phi - hepatocyte.sinusoid_mean.phi)) <= sin(pi/4) ? 1 : 0) : (abs(sin(membrane.phi - hepatocyte.sinusoid_mean.phi)) <= sin(pi/4) ? 1 : 0)</Expression>
</Equation>
<!-- <Disabled>
<Constant symbol="hepatocyte.proliferation_p" name="division rate from Fig.14 in Hoehme et al. 2018: 1/(1080min/(5min/timestep))=1/(216 time steps)" tags="HCC" value="1/216"/>
</Disabled>
-->
<!-- <Disabled>
<System time-step="12" solver="Euler [fixed, O(1)]" tags="CCl4">
<Rule symbol-ref="hepatocyte.proliferation_p">
<Annotation>Definition of the cell division probability of BrdU-positive hepatocytes depending on the the layer number.</Annotation>
<Expression>if(time.regeneration >= 2.9 - hepatocyte.time_span.proliferation/2 && time.regeneration < 2.9 + hepatocyte.time_span.proliferation/2,
if(hepatocyte.layer.self == 1, 0.3,
if(hepatocyte.layer.self == 2, 0.25,
if(hepatocyte.layer.self == 3, 0.2,
if(hepatocyte.layer.self == 4, 0.2, 0.1)))), 0)</Expression>
</Rule>
</System>
</Disabled>
-->
<Property symbol="hepatocyte.time.division" tags="HCC" value="hepatocyte.time_span.cell_cycle"/>
<Property symbol="hepatocyte.time.nextdivision" tags="HCC" value="rand_norm(hepatocyte.time_span.cell_cycle,hepatocyte.time_span.cell_cycle_delta)"/>
<CellDivision trigger="on-change" division-plane="random" name="proliferation w/o HSA" tags="HCC">
<Condition>time.days >= hepatocyte.time.birth+hepatocyte.time.division</Condition>
<Triggers>
<Rule symbol-ref="hepatocyte.time.birth">
<Expression>time.days</Expression>
</Rule>
<Rule symbol-ref="hepatocyte.time.nextdivision" tags="HCC">
<Expression>rand_norm(hepatocyte.time_span.cell_cycle,hepatocyte.time_span.cell_cycle_delta)</Expression>
</Rule>
</Triggers>
</CellDivision>
<!-- <Disabled>
<Event trigger="when-true" name="truncate Gaussian for individual cell cycle duration" tags="HCC">
<Condition>time.days < hepatocyte.time.birth+2*mcs_duration</Condition>
<Rule symbol-ref="hepatocyte.time.division" tags="HCC">
<Expression>if( hepatocyte.time.nextdivision < (hepatocyte.time_span.cell_cycle-hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle-hepatocyte.time_span.cell_cycle_delta), if( hepatocyte.time.nextdivision > (hepatocyte.time_span.cell_cycle+hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle+hepatocyte.time_span.cell_cycle_delta), hepatocyte.time.nextdivision))</Expression>
</Rule>
</Event>
</Disabled>
-->
<!-- <Disabled>
<Chemotaxis strength="100" tags="CCl4, HCC" field="c"/>
</Disabled>
-->
<System time-step="mcs_duration" name="truncate Gaussian for individual cell cycle duration" solver="Euler [fixed, O(1)]" tags="HCC">
<Rule symbol-ref="hepatocyte.time.division" tags="HCC">
<Expression>if(hepatocyte.time.nextdivision < (hepatocyte.time_span.cell_cycle - hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle - hepatocyte.time_span.cell_cycle_delta),
if(hepatocyte.time.nextdivision > (hepatocyte.time_span.cell_cycle + hepatocyte.time_span.cell_cycle_delta), (hepatocyte.time_span.cell_cycle + hepatocyte.time_span.cell_cycle_delta), hepatocyte.time.nextdivision))</Expression>
</Rule>
</System>
</CellType>
</CellTypes>
<CPM>
<Interaction default="0.0">
<Contact type1="hepatocytes" type2="medium" value="0"/>
<Contact type1="hepatocytes" type2="hepatocytes" value="hepatocyte.contactEnergy">
<HomophilicAdhesion strength="hepatocyte.adhesionStrength" adhesive="membrane.apical" tags="CCl4, HCC"/>
</Contact>
<Contact type1="hepatocytes" type2="CV" value="hepatocyte.contactEnergy"/>
<Contact type1="hepatocytes" type2="PV" value="hepatocyte.contactEnergy"/>
<Contact type1="HCC_with_HSA" type2="HCC_with_HSA" value="hepatocyte.contactEnergy">
<HomophilicAdhesion strength="hepatocyte.adhesionStrength" adhesive="membrane.apical" tags="HCC"/>
</Contact>
<Contact type1="HCC_with_HSA" type2="CV" value="hepatocyte.contactEnergy"/>
<Contact type1="HCC_with_HSA" type2="PV" value="hepatocyte.contactEnergy"/>
<Contact type1="HCC_without_HSA" type2="HCC_without_HSA" value="hepatocyte.contactEnergy">
<HomophilicAdhesion strength="hepatocyte.adhesionStrength" adhesive="membrane.apical" tags="HCC"/>
</Contact>
<Contact type1="HCC_without_HSA" type2="CV" value="hepatocyte.contactEnergy"/>
<Contact type1="HCC_without_HSA" type2="PV" value="hepatocyte.contactEnergy"/>
<Contact type1="hepatocytes" type2="HCC_with_HSA" value="hepatocyte.contactEnergy">
<HomophilicAdhesion strength="hepatocyte.adhesionStrength" adhesive="membrane.apical" tags="HCC"/>
</Contact>
<Contact type1="hepatocytes" type2="HCC_without_HSA" value="hepatocyte.contactEnergy">
<HomophilicAdhesion strength="hepatocyte.adhesionStrength" adhesive="membrane.apical" tags="HCC"/>
</Contact>
<Contact type1="HCC_with_HSA" type2="HCC_without_HSA" value="hepatocyte.contactEnergy">
<HomophilicAdhesion strength="hepatocyte.adhesionStrength" adhesive="membrane.apical" tags="HCC"/>
</Contact>
</Interaction>
<MonteCarloSampler stepper="edgelist">
<MCSDuration symbol="mcs_duration" value="3.0"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<MetropolisKinetics temperature="0.4"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>3</Order>
</Neighborhood>
</ShapeSurface>
</CPM>
<Analysis>
<ModelGraph format="svg" reduced="false" include-tags="#untagged,CCl4,HCC"/>
<Gnuplotter time-step="144" decorate="true" tags="CCl4">
<Annotation>plot at least every 144 time steps = 0.5d to select plots for 1008=3d, 1584=5d, 2016=6.5d as in Fig.3</Annotation>
<Terminal name="png" size="1534, 1200, 0"/>
<Plot>
<Cells value="cell.type">
<ColorMap adaptive-range="false">
<Color value="0" color="white"/>
<Color value="1" color="red"/>
<Color value="2" color="blue"/>
<Color value="3" color="light-salmon"/>
<Color value="4" color="mediumpurple3"/>
<Color value="5" color="forest-green"/>
</ColorMap>
</Cells>
</Plot>
<!-- <Disabled>
<Plot title="Apical sides">
<Cells opacity="0.1" flooding="true" value="membrane.apical">
<ColorMap>
<Color value="0" color="gray90"/>
<Color value="1" color="light-green"/>
</ColorMap>
</Cells>
</Plot>
</Disabled>
-->
<!-- <Disabled>
<Plot>
<Field symbol-ref="c"/>
</Plot>
</Disabled>
-->
</Gnuplotter>
<Logger time-step="6" name="log clone shape every 30min" tags="HCC">
<Input>
<Symbol symbol-ref="time.post_init"/>
<Symbol symbol-ref="celltype.HCC_with_HSA.size"/>
<Symbol symbol-ref="clone.withHSA.aspectratio"/>
<!-- <Disabled>
<Symbol symbol-ref="data.withHSA.mse"/>
</Disabled>
-->
<Symbol symbol-ref="celltype.HCC_without_HSA.size"/>
<Symbol symbol-ref="clone.withoutHSA.aspectratio"/>
<!-- <Disabled>
<Symbol symbol-ref="data.withoutHSA.mse"/>
</Disabled>
-->
</Input>
<Output>
<TextOutput/>
</Output>
<Plots>
<Plot time-step="-1" title="Tumor growth kinetics">
<Style style="points"/>
<Terminal terminal="png"/>
<X-axis minimum="0.0">
<Symbol symbol-ref="time.post_init"/>
</X-axis>
<Y-axis minimum="1" maximum="1000" logarithmic="true">
<Symbol symbol-ref="celltype.HCC_with_HSA.size"/>
<Symbol symbol-ref="celltype.HCC_without_HSA.size"/>
</Y-axis>
</Plot>
<Plot time-step="-1" title="Tumor shape time course">
<Style style="linespoints"/>
<Terminal terminal="png"/>
<X-axis>
<Symbol symbol-ref="time.post_init"/>
</X-axis>
<Y-axis>
<Symbol symbol-ref="clone.withHSA.aspectratio"/>
<Symbol symbol-ref="clone.withoutHSA.aspectratio"/>
</Y-axis>
</Plot>
<Plot time-step="-1" title="Tumor shape characteristics with HSA">
<Style style="lines"/>
<Terminal terminal="png"/>
<X-axis minimum="4" maximum="100" logarithmic="true">
<Symbol symbol-ref="celltype.HCC_with_HSA.size"/>
</X-axis>
<Y-axis minimum="1.0" maximum="3.5">
<Symbol symbol-ref="clone.withHSA.aspectratio"/>
<Symbol symbol-ref="data.withHSA.max"/>
<Symbol symbol-ref="data.withHSA.min"/>
</Y-axis>
</Plot>
<Plot time-step="-1" title="Tumor shape characteristics without HSA">
<Style style="lines"/>
<Terminal terminal="png"/>
<X-axis minimum="4" maximum="100" logarithmic="true">
<Symbol symbol-ref="celltype.HCC_without_HSA.size"/>
</X-axis>
<Y-axis minimum="1.0" maximum="3.5">
<Symbol symbol-ref="clone.withoutHSA.aspectratio"/>
<Symbol symbol-ref="data.withoutHSA.max"/>
<Symbol symbol-ref="data.withoutHSA.min"/>
</Y-axis>
</Plot>
</Plots>
</Logger>
<Function symbol="data.withHSA.max" name="Data (upper limit)" tags="HCC">
<Expression>-0.326*ln(celltype.HCC_with_HSA.size)+3.55</Expression>
</Function>
<Function symbol="data.withHSA.min" name="Data (lower limit)" tags="HCC">
<Expression>-0.195*ln(celltype.HCC_with_HSA.size)+2.44</Expression>
</Function>
<Function symbol="data.withoutHSA.max" name="Data (upper limit)" tags="HCC">
<Expression>-0.130*ln(celltype.HCC_without_HSA.size)+2.28</Expression>
</Function>
<Function symbol="data.withoutHSA.min" name="Data (lower limit)" tags="HCC">
<Expression>-0.029*ln(celltype.HCC_without_HSA.size)+1.24</Expression>
</Function>
</Analysis>
<CellPopulations>
<Population type="hepatocytes" name="Hepatocytes" size="0">
<InitCircle name="Hepatocyte initialization" number-of-cells="500" tags="CCl4, HCC" mode="random">
<Annotation>Hepatocyte density (lit.): 1889 cells/mm^2
Lobule area in 2D slice (lit.): 0.21 mm^2
- Hepatocyte lesion without hepatocytes, i.e. with medium only (model): 0.2043646748 mm^2 → 386 cells
- Lobule area, i.e. includung medium, hepatocytes, CV, PVs (model): 0.2132578897 mm^2 → 403 cells</Annotation>
<Dimensions radius="size.x/2" center="size.x/2, size.y/2 * sqrt(3)/2, 0"/>
</InitCircle>
</Population>
<Population type="CV" name="Central vein" size="1">
<InitCellObjects name="Set coordinates" tags="CCl4, HCC" mode="order">
<Annotation>Central vein radius (lit.): 41.2 µm</Annotation>
<Arrangement displacements="1, 1, 1" repetitions="1, 1, 1">
<Sphere radius="41.2/node.length" center="size.physical/2"/>
</Arrangement>
</InitCellObjects>
</Population>
<Population type="PV" name="Portal vein" size="1">
<InitCellObjects name="bottom left" tags="CCl4, HCC" mode="order">
<Arrangement displacements="1, 1, 1" repetitions="1, 1, 1">
<Sphere radius="23.3 / node.length" center="52.5, 5 * sqrt(3)/2, 0"/>
</Arrangement>
</InitCellObjects>
<InitCellObjects name="bottom right" tags="CCl4, HCC" mode="order">
<Arrangement displacements="1, 1, 1" repetitions="1, 1, 1">
<Sphere radius="23.3 / node.length" center="147.5, 5 * sqrt(3)/2, 0"/>
</Arrangement>
</InitCellObjects>
<InitCellObjects name="mid left" tags="CCl4, HCC" mode="order">
<Arrangement displacements="1, 1, 1" repetitions="1, 1, 1">
<Sphere radius="23.3 / node.length" center="5, 100 * sqrt(3)/2, 0"/>
</Arrangement>
</InitCellObjects>
<InitCellObjects name="mid right" tags="CCl4, HCC" mode="order">
<Arrangement displacements="1, 1, 1" repetitions="1, 1, 1">
<Sphere radius="23.3 / node.length" center="195, 100 * sqrt(3)/2, 0"/>
</Arrangement>
</InitCellObjects>
<InitCellObjects name="upper left" tags="CCl4, HCC" mode="order">
<Arrangement displacements="1, 1, 1" repetitions="1, 1, 1">
<Sphere radius="23.3 / node.length" center="52.5, 195 * sqrt(3)/2, 0"/>
</Arrangement>
</InitCellObjects>
<InitCellObjects name="upper right" tags="CCl4, HCC" mode="order">
<Arrangement displacements="1, 1, 1" repetitions="1, 1, 1">
<Sphere radius="23.3 / node.length" center="147.5, 195 * sqrt(3)/2, 0"/>
</Arrangement>
</InitCellObjects>
</Population>
<Population type="HCC_with_HSA" name="HCC with HSA" size="1">
<InitCellObjects tags="HCC" mode="distance">
<Arrangement displacements="60, 60, 1" repetitions="1, 1, 1">
<Sphere radius="2.0" center="size.x/2-30, size.y/2-45, 0.0"/>
</Arrangement>
</InitCellObjects>
</Population>
<Population type="HCC_without_HSA" name="HCC without HSA" size="1">
<InitCellObjects tags="HCC" mode="distance">
<Arrangement displacements="60, 60, 1" repetitions="1, 1, 1">
<Sphere radius="2.0" center="size.x/2+22, size.y/2+27, 0.0"/>
</Arrangement>
</InitCellObjects>
</Population>
</CellPopulations>
</MorpheusModel>

Downloads
Files associated with this model:
hoehme-2018_fig-14_tumor-growth-kinetics.jpghoehme-2018_fig-3_tumor-growth-scenarios.jpghoehme-2018_fig-4_tumor-axis-ratios.jpghoehme-2018_fig-5_rat-liver.jpgM9148_hcc_fig-4_aspect-ratio@2x.jpgM9148_hcc_fig-5_tumor-growth-kinetics.pngM9148_hcc_fig-6_tumor-growth-kinetics.pngM9148_hcc_model-graph.svgM9148_hcc_model.xmlM9148_hcc_vid-1_simulation.mp4