Simulation-Based Inference of Cell Migration Dynamics in Complex Spatial Environments
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Simulating cell migration within an environment of structural obstacles
Introduction
This repository contains the Morpheus model configuration file for simulating cell migration within an environment of structural obstacles.
The model is built to explore how cell populations respond to chemokine gradients and physical boundaries, using a Cellular Potts Model (CPM) and persistent motion dynamics.
Description
- 2D Cellular Potts Model
- Chemokine Gradient: Gaussian profile centered around a virtual source
- Initial Cell Distribution: Random circular placement near one-third of the grid’s height
- Motion Behavior:
- Persistent random walk with exponential waiting times (tumbling)
- Chemotaxis toward a static Gaussian chemokine field
- Structural obstacles are read from a TIFF image mask
- Logging of cell position every 30 time steps
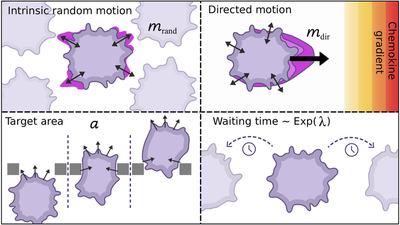
Results
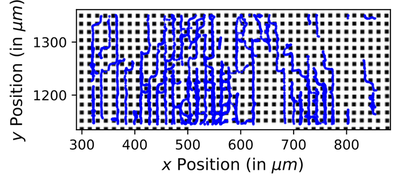
We integrated experimental observations of dendritic cell migration in a geometrically constrained microenvironment with a calibrated Cellular Potts Model and demonstrate that these spatial constraints modulate motility dynamics, including velocity and directional changes. We show that classical summary statistics used in approximate Bayesian computation (ABC)—such as mean squared displacement and turning angle distributions—can resolve key mechanistic features, however fail to extract richer spatiotemporal patterns. Instead, we apply neural posterior estimation (NPE) with neural network based summary features, which outperform handcrafted statistics. This learned summary representation of the data enables robust and flexible parameter inference, providing a data-driven framework for model calibration and advancing the quantitative analysis of cell migration in structured microenvironments.
Reference
This model is the original used in the publication, up to technical updates:
J. Arruda, E. Alamoudi, R. Müller, M. Vaisband, R. Molkenbur, J. Merrin, E. Kiermaier, J. Hasenauer: Simulation-based inference of cell migration dynamics in complex spatial environments. npj Syst. Biol. Appl. 12 (20), 2026.
Model
cell_movement_v24.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="4">
<Description>
<Details>Model ID: https://identifiers.org/morpheus/M6342
Software: Morpheus (open source). Download from: https://morpheus.gitlab.io
Full title: Simulation-Based Inference of Cell Migration Dynamics in Complex Spatial Environments
Authors: J. Arruda, E. Alamoudi, R. Müller, M. Vaisband, R. Molkenbur, J. Merrin, E. Kiermaier, J. Hasenauer
Submitter: J. Arruda
Curator: D. Jahn
Date: 14.07.2025
Reference: This model is the original used in the publication, up to technical updates:
J. Arruda, E. Alamoudi, R. Müller, M. Vaisband, R. Molkenbur, J. Merrin, E. Kiermaier, J. Hasenauer: Simulation-based inference of cell migration dynamics in complex spatial environments. npj Syst. Biol. Appl. 12 (20), 2026.
https://doi.org/10.1038/s41540-026-00648-9
Comment: Cell migration in a complex spatial environment.</Details>
<Title>Cell_movement_grid_v24</Title>
</Description>
<Space>
<Lattice class="square">
<Size symbol="size" value="1173,2500,0"/>
<BoundaryConditions>
<Condition type="constant" boundary="x"/>
<Condition type="constant" boundary="y"/>
<Condition type="constant" boundary="-x"/>
<Condition type="constant" boundary="-y"/>
</BoundaryConditions>
<Neighborhood>
<Distance>2.0</Distance>
</Neighborhood>
<Domain boundary-type="noflux">
<Image path="Cell_migration_grid_v3_final2_invers.tiff"/>
</Domain>
</Lattice>
<SpaceSymbol symbol="space"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="60000"/>
<!-- <Disabled>
<StopTime value="5000"/>
</Disabled>
-->
<TimeSymbol symbol="time"/>
<SaveInterval value="0"/>
<!-- <Disabled>
<RandomSeed value="0"/>
</Disabled>
-->
</Time>
<Analysis>
<ModelGraph format="dot" reduced="false" include-tags="#untagged"/>
<Logger time-step="30">
<Input>
<Symbol symbol-ref="nodeL"/>
<Symbol symbol-ref="cell.volume"/>
<Symbol symbol-ref="cell.center.x"/>
<Symbol symbol-ref="cell.center.y"/>
</Input>
<Output>
<TextOutput/>
</Output>
</Logger>
<!-- <Disabled>
<Gnuplotter time-step="500">
<Plot>
<Field symbol-ref="u1"/>
<Cells internal:disabled="true" value="cell.id"/>
</Plot>
<Terminal name="png"/>
<Plot>
<Field symbol-ref="dummy"/>
<Cells value="cell.id"/>
</Plot>
<Plot>
<Cells value="cell.id"/>
<Field symbol-ref="u1"/>
<CellArrows orientation="5 * move_dir"/>
</Plot>
</Gnuplotter>
</Disabled>
-->
</Analysis>
<CPM>
<Interaction>
<Contact value="40" type1="cells" type2="cells"/>
<Contact value="20" type1="cells" type2="medium"/>
<Contact value="1000" type1="cells" type2="boundary"/>
</Interaction>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<MetropolisKinetics yield="0.05" temperature="6"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>4</Order>
</Neighborhood>
</ShapeSurface>
</CPM>
<Global>
<Constant symbol="tumble.run_duration" value="0"/>
<Constant symbol="cell_nodes" name="CPM cell volume (μm^2 / 1.31^2)" value="cell_nodes_real / (nodeL*nodeL)"/>
<Constant symbol="cell_nodes_real" name="CPM cell volume μm^2" value="143"/>
<Constant symbol="x0_chemokine" name="position chemokine source in um" value="x0_sim * nodeL"/>
<Constant symbol="y0_chemokine" name="position chemokine source in μm" value="hole.diam+hole.diam/2+270"/>
<Field symbol="dummy" value="-1"/>
<Constant symbol="x0_sim" name="position chemokine source for simulation" value="size.x/2"/>
<Constant symbol="y0_sim" name="position chemokine source for simulation" value="y0_chemokine/nodeL"/>
<Constant symbol="sigma0" name="spread of inital concentration (std)" value="550"/>
<Constant symbol="nodeL" name="factor from simulation lattice ength to μm" value="1.31"/>
<Constant symbol="peak_concentration" name="peak concentration of the chemokine (total of 7ng mass of the chemokine)" value="7e+6"/>
<Field symbol="u1" value="peak_concentration/(2*pi*(sigma0^2))*exp(-1/2*(((space.x)-(x0_sim))^2+ ((space.y)-(y0_sim))^2)/(sigma0^2)) "/>
<Constant symbol="num_cells" name="number of cell in simulation" value="143"/>
<Constant symbol="hole.diam" name="diameter of the hole in μm" value="1500"/>
<Constant symbol="hole.diam_sim" name="diameter of the hole in simulation" value="hole.diam/nodeL"/>
</Global>
<CellPopulations>
<Population type="cells" size="0">
<InitCircle name="Starting area of the cells" number-of-cells="num_cells" mode="random">
<Dimensions radius="65 / nodeL" center="size.x/2,size.y/3,0"/>
</InitCircle>
</Population>
<BoundaryValue boundary="domain" value="boundary"/>
</CellPopulations>
<CellTypes>
<CellType class="medium" name="medium">
<Property symbol="l_U" value="0"/>
</CellType>
<CellType class="medium" name="boundary"/>
<CellType class="biological" name="cells">
<VolumeConstraint name="target volume in μm^2 / (nodeL*nodeL)" strength="1" target="cell_nodes"/>
<Constant symbol="alpha_start" name="angle for random starting direction" value="rand_uni(0,2*pi) "/>
<PropertyVector symbol="move.dir" name="random direction" value="cos(alpha_start), sin(alpha_start), 0 "/>
<Constant symbol="move.duration.scale" name="Mean of exponential distribution (seconds)" value="1 / move.duration.rate"/>
<Constant symbol="move.duration.shape" name="shape of gamma distribution fixed to be an exponential" value="1"/>
<Constant symbol="move.strength" name="strength of directed movement" value="10"/>
<Constant symbol="move.duration.rate" name="Rate of exponential waiting time (1/seconds)" value="0.1 "/>
<Property symbol="tumble.run_duration" name="run duration" value="0.0"/>
<Property symbol="tumble.last" name="last tumble event" value="0"/>
<DirectedMotion name="Persistent.Random.Walk" strength="move.strength" direction="move.dir"/>
<Event name="tumble" trigger="when-true">
<Condition>time >= tumble.last + tumble.run_duration</Condition>
<Rule symbol-ref="tumble.last" name="last tumble event">
<Expression>time</Expression>
</Rule>
<Rule symbol-ref="tumble.run_duration" name="new update time (gamma distributed)">
<Expression>rand_gamma(move.duration.shape, move.duration.scale) </Expression>
</Rule>
<Intermediate symbol="alpha" name="angle for random direction" value="rand_uni(0,2*pi)"/>
<VectorRule symbol-ref="move.dir" notation="x,y,z" name="random direction">
<Expression>cos(alpha), sin(alpha) , 0</Expression>
</VectorRule>
</Event>
<Constant symbol="gradient_strength" name="gradient of chemokine" value="1"/>
<Chemotaxis strength="gradient_strength*1000" field="u1"/>
</CellType>
</CellTypes>
</MorpheusModel>

Downloads
Files associated with this model: