Human Airway Infection
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Viral Spread within the Human Airway Epithelium (HAE)
Introduction
The spread of viral infections within tissues depends on several factors including the spatial distribution and turnover dynamics of target cells. In Raach et al. (2023), a Cellular Potts model was developed to investigate the influence of target cell heterogeneity within the human airway epithelium on SARS-CoV-2 infection dynamics. The study aims to understand how different tissue compositions and regenerative turnover throughout the respiratory tree affect the progression of SARS-CoV-2 infections. The model incorporates cell-type specific infection kinetics, tissue regeneration, and accounts for both global and local spread of infections through cell-free and cell-associated viral transmission mechanisms.
Parameterization of the model is achieved by integrating published experimental data on differentiating primary human bronchial epithelial cells within air-liquid interface cultures through approximate Bayesian computation (de Borja Callejas et al., 2014, Schamberger et al., 2015). By simulating SARS-CoV-2 infections given various tissue compositions and regenerative capacities, the model provides insights into cell-type specific infection dynamics and the impact of tissue composition on disease progression and pathology.
Description
The human airway epithelium (HAE) cell culture system is represented by a 2D individual cell-based model within a radial lattice grid. The simulation initializes a total population of $\approx 4.7 \times 10^4$ cells, with each cell occupying a target area of 60 lattice sites. The model considers three different cell types: basal (B), secretory (S), and ciliated (C) cells, which constitute the epithelium with different relative contributions. To mimic the pseudostratified 3D-morphology of the upper respiratory epithelium, the volume strength of basal cells is set to $\frac{1}{10}$ of that of secretory or ciliated cells, allowing basal cells to be ‘overgrown’ by the other cell types, i.e., being potentially compressed to only one lattice site in fully differentiated tissue.
Cellular turnover and epithelial regeneration follows a linear differentiation process with continuous proliferation of basal cells at a density-dependent rate ($\alpha$), subsequent differentiation into secretory and ciliated cells ($\lambda_\mathrm{S}$, $\lambda_\mathrm{C}$) and cell loss according to cell-type specific death rates ($\delta_\mathrm{B}$, $\delta_\mathrm{S}$, $\delta_\mathrm{C}$) (see also Fig. 1). The processes of cell proliferation, differentiation, and cell death are incorporated into the Morpheus model by translating the model system shown in Fig. 1 into a probabilistic framework (see also Raach et al., 2023).
: [**Raach _et al._, 2023**](#reference), [Fig. 1B, D](https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1011356#pcbi-1011356-g001).)](/media/model/m6296/M6296_hu3a9d5e3cfb4fd4e38126162b6a8d309f_780463_3c4844592793f7f2b3b19ce2f28d491b.png)
To incorporate viral infection, the model follows the standard model of viral dynamics: After infection of a cell, the cell progresses through an eclipse phase before it becomes infectious and starts to produce new virions. Each cell type can get infected through either cell-free or cell-associated transmission modes dependent on the viral concentration or the number of infectious cells in its neighborhood, respectively, as well as the cell-type specific transmission rates, $\beta_\mathrm{cf}$ and $\beta_\mathrm{cc}$ (see also Raach et al., 2023 for further details).
The model code M6296_human-airway-infection_model.xml provided here simulates viral infection within bronchial epithelium given an initial cell type distribution of $39.7\ \text{%}$ (B), $13.4\ \text{%}$ (S), and $46.9\ \text{%}$ (C), at equilibrium before infection. A simulation time step represents $0.25$ hours in real time. Infection is initialized by adding a viral load of $2.6 \times 10^6$ RNA copies at $6.25$ hours ($= 25$ time steps) post simulation start, with the epithelium being at confluence. For all individual parameters, see also Raach et al., 2023.
Results
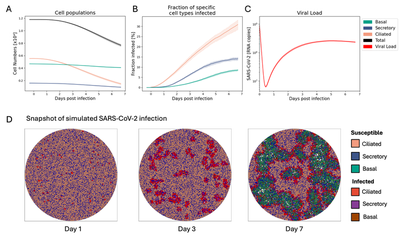
Vid. 1 and Fig. 2 show the results of re-running the model with the parametrizations and initial cell distributions as outlined in Raach et al., 2023 over a time period of $7$ days.
Results reproduce the findings reported by Raach et al. with ciliated cells showing the highest fraction of infected cells (Fig. 2B). Snapshots of the Cellular Potts model (Fig. 2D) indicate the effect of heterogeneous infection patterns on tissue integrity (see also Raach et al., 2023).
Reference
This model is the original used in the publication, up to technical updates:
B. Raach, N. Bundgaard, M. J. Haase, J. Starruß, R. Sotillo, M. L. Stanifer, F. Graw: Influence of cell type specific infectivity and tissue composition on SARS-CoV-2 infection dynamics within human airway epitheliumn. PLoS Comput. Biol. 19 (8): e1011356, 2023.
Model
M6296_human-airway-infection_model.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="4">
<Description>
<Details>Model ID: https://identifiers.org/morpheus/M6296
Title: Human Airway Infection
Authors: B. Raach, N. Bundgaard, M. J. Haase, J. Starruß, R. Sotillo, M. L. Stanifer, F. Graw
Submitters: P. Lukas, F. Graw
Curators: D. Jahn
Date: 08.05.2024
Software: Morpheus (open source). Download from: https://morpheus.gitlab.io
Units: [time] = 0.25 h
[space] = 1 µm
Reference: This model is the original used in the publication, up to technical updates:
B. Raach, N. Bundgaard, M. J. Haase, J. Starruß, R. Sotillo, M. L. Stanifer, F. Graw: Influence of cell type specific
infectivity and tissue composition on SARS-CoV-2 infection dynamics within human airway epitheliumn. PLoS
Comput. Biol. 19 (8): e1011356, 2023.
https://doi.org/10.1371/journal.pcbi.1011356</Details>
<Title>M6296 Human Airway Infection</Title>
</Description>
<Space>
<Lattice class="square">
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<Size symbol="size" value="simulation_diameter,simulation_diameter,0"/>
<BoundaryConditions>
<Condition type="noflux" boundary="x"/>
<Condition type="noflux" boundary="y"/>
<!-- <Disabled>
<Condition type="noflux" boundary="-x"/>
</Disabled>
-->
<!-- <Disabled>
<Condition type="noflux" boundary="-y"/>
</Disabled>
-->
</BoundaryConditions>
<Domain boundary-type="noflux">
<Circle diameter="1204"/>
</Domain>
</Lattice>
<SpaceSymbol symbol="space"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="500"/>
<TimeSymbol symbol="time"/>
</Time>
<Analysis>
<ModelGraph include-tags="#untagged" format="png" reduced="false"/>
<Gnuplotter time-step="96" decorate="false">
<Plot>
<Cells value="cell.type">
<ColorMap adaptive-range="false">
<Color color="sea-green" value="1"/>
<Color color="brown4" value="2"/>
<Color color="brown4" value="3"/>
<Color color="medium-blue" value="4"/>
<Color color="dark-violet" value="5"/>
<Color color="dark-violet" value="6"/>
<Color color="dark-salmon" value="7"/>
<Color color="red" value="8"/>
<Color color="red" value="9"/>
<Color color="light-green" value="10"/>
<Color color="light-blue" value="11"/>
<Color color="gold" value="12"/>
<Color color="black" value="13"/>
</ColorMap>
</Cells>
</Plot>
<Terminal name="png" size="2000, 2000, 0"/>
</Gnuplotter>
</Analysis>
<Global>
<Constant symbol="simulation_diameter" value="1204">
<Annotation>IF CHANGED: also has to be manually changed under "Space"-"Lattice"-"Domain"-"Circle"-"diameter"</Annotation>
</Constant>
<Constant symbol="Nmax" value="47177*1.288">
<Annotation>Maximum cell capacity based on ALI-culture experiment (47177 cells) scaled by 1.288 to reproduce seeding conditions reported by Raach et al.</Annotation>
</Constant>
<Constant symbol="timestepsize" value="0.25">
<Annotation>Time resolution of 0.25 hours per computational step (4 steps / h) to capture rapid viral and cellular events.</Annotation>
</Constant>
<Constant symbol="init_cell_number" value="47177">
<Annotation>Initial epithelial cell density measured in vitro and used to seed the simulation lattice.</Annotation>
</Constant>
<Constant symbol="alphaB" value="0.338/24*timestepsize">
<Annotation>Basal division parameter: 0.338 divisions / day (Fig 2, Raach et al.) converted to per-timestep probability.</Annotation>
</Constant>
<Constant symbol="diffS" value="0.084/24*timestepsize*4">
<Annotation>Secretory differentiation rate: 0.084 per day (Fig 3a) scaled to timestep and cell cycle phases (4 subdivisions).</Annotation>
</Constant>
<Constant symbol="diffCil" value="0.162/24*timestepsize*4">
<Annotation>Ciliated differentiation rate: 0.162 per day (Fig 3a) converted to per-step based on 4-phase progression.</Annotation>
</Constant>
<Constant symbol="deltaB" value="0.0139/24*timestepsize">
<Annotation>Basal homeostatic death rate of 0.0139 / day (Raach et al.), normalized to simulation time steps.</Annotation>
</Constant>
<Constant symbol="deltaS" value="0.083/24*timestepsize">
<Annotation>Secretory cell death rate: 0.083 / day (Fig 3b) converted to per-step probability.</Annotation>
</Constant>
<Constant symbol="deltaCil" value="0.029/24*timestepsize
">
<Annotation>Ciliated cell death rate of 0.029 / day (Fig 3b) converted for 0.25 h steps.</Annotation>
</Constant>
<Function symbol="cell_number">
<Expression>sum(celltype.secretory.size, celltype.secretory_infected.size, celltype.secretory_infectious.size ,celltype.ciliated.size, celltype.ciliated_infected.size, celltype.ciliated_infectious.size, celltype.basal.size, celltype.basal_infected.size, celltype.basal_infectious.size)</Expression>
<Annotation>Aggregates total live cell count across all types to calculate density-dependent processes.</Annotation>
</Function>
<Constant symbol="locality" value="15">
<Annotation>Spatial radius (in lattice units) for local neighborhood infection interactions.</Annotation>
</Constant>
<Constant symbol="betaB" value="10^(-9.367795)/locality*10000*timestepsize/3*(1-cc_ct_ratio)">
<Annotation>Basal cell infection coefficient: calibrated from experimental MOI and adjusted for local viral concentration and cell-cell transmission factors.</Annotation>
</Constant>
<Constant symbol="betaS" value="10^(-9.403180)/locality*10000*timestepsize/3*(1-cc_ct_ratio)">
<Annotation>Secretory cell infection rate: reflects ACE2 expression levels with adjustments for CC interactions.</Annotation>
</Constant>
<Constant symbol="betaC" value="10^(-9.430063)*10000/locality*timestepsize*(1-cc_ct_ratio)">
<Annotation>Ciliated cell infection parameter: highest susceptibility, derived from fitted infection curves.</Annotation>
</Constant>
<Constant symbol="kappaB" value="4*0.192848*timestepsize">
<Annotation>Basal eclipse-to-infectious progression rate: 0.1928 / day, four internal compartments per cell cycle.</Annotation>
</Constant>
<Constant symbol="kappaS" value="4*0.182802*timestepsize">
<Annotation>Secretory eclipse progression: 0.1828 / day scaled for four internal compartments per cell cycle</Annotation>
</Constant>
<Constant symbol="kappaC" value="4*0.217471*timestepsize">
<Annotation>Ciliated eclipse progression constant (0.2175 / day)scaled for four internal compartments per cell cycle.</Annotation>
</Constant>
<Constant symbol="rhoB" value="10^(4.376343)*timestepsize">
<Annotation>Basal viral secretion rate: 10^4.376 units / h based on in vitro output, scaled by timestep.</Annotation>
</Constant>
<Constant symbol="rhoS" value="10^(4.613919)*timestepsize">
<Annotation>Secretory cells produce 10^4.614 virions / h in HAE cultures (Raach et al.), normalized per-step.</Annotation>
</Constant>
<Constant symbol="rhoC" value="10^(4.689795)*timestepsize">
<Annotation>Ciliated cell secretion rate 10^4.690 / h, reflecting highest viral output per cell.</Annotation>
</Constant>
<Constant symbol="db_I" value="4*0.080933*timestepsize">
<Annotation>Basal infectious death rate: 0.0809 / day </Annotation>
</Constant>
<Constant symbol="ds_I" value="4*0.077071*timestepsize">
<Annotation>Secretory infectious death parameter: 0.0771 / day</Annotation>
</Constant>
<Constant symbol="dc_I" value="4*0.074392*timestepsize">
<Annotation>Ciliated infectious death rate: 0.0744 / day</Annotation>
</Constant>
<Constant symbol="c" value="0.63*timestepsize">
<Annotation>Extracellular viral clearance rate of 0.63 / h (decay constant), scaled to simulation time step.</Annotation>
</Constant>
<Field symbol="ViralLoad" value="0.0">
<Diffusion rate="20"/>
<Annotation>Extracellular viral concentration field; diffusion coefficient 20 µm² / h</Annotation>
</Field>
<Variable symbol="add_infection" value="0"/>
<System time-step="1.0" solver="Bogacki-Shampine [adaptive, O(3)]">
<DiffEqn symbol-ref="ViralLoad">
<Expression>viralsecretion + add_infection - c * ViralLoad</Expression>
</DiffEqn>
<Annotation>Extracellular viral concentration field; diffusion coefficient 20 µm² / h</Annotation>
</System>
<Event trigger="when-true">
<Condition>time >= 25</Condition>
<Rule symbol-ref="add_infection">
<Expression>0</Expression>
</Rule>
</Event>
<Event>
<Condition>time >= 24.5 and time < 25.5</Condition>
<Rule symbol-ref="add_infection">
<Expression>2.6*10^6/(3.14/4*simulation_diameter^2)*init_cell_number/47177*800</Expression>
</Rule>
<Annotation>add same single-cell viral load per cell as in ali-culture experiment </Annotation>
</Event>
<Variable symbol="total_viral_load" value="0.0"/>
<Mapper>
<Input value="ViralLoad"/>
<Output symbol-ref="total_viral_load" mapping="sum"/>
</Mapper>
<Constant symbol="ccbetaB" value="1.0*cc_reduced*cc_ct_ratio/cceffectivness">
<Annotation>Cell–cell transmission factor for basal cells, scaled by effectiveness and CT ratio.</Annotation>
</Constant>
<Constant symbol="ccbetaS" value="1.536*cc_reduced*cc_ct_ratio/cceffectivness">
<Annotation>Cell–cell transmission factor for secretorty cells, scaled by effectiveness and CT ratio.</Annotation>
</Constant>
<Constant symbol="ccbetaC" value="4.33*cc_ct_ratio/cceffectivness">
<Annotation>Cell–cell transmission factor for cilitaed cells, scaled by effectiveness and CT ratio.</Annotation>
</Constant>
<Constant symbol="ccrhoB" value="1*timestepsize">
<Annotation>Basal cell per-step contribution to neighbor infection strength from empirical fit.</Annotation>
</Constant>
<Constant symbol="ccrhoS" value="0.5786*timestepsize">
<Annotation>Secretory cell per-step contribution to neighbor infection strength from empirical fit.</Annotation>
</Constant>
<Constant symbol="ccrhoC" value="0.48590*timestepsize">
<Annotation>Ciliated cell per-step contribution to neighbor infection strength from empirical fit.</Annotation>
</Constant>
<Constant symbol="cceffectivness" value="20">
<Annotation>Normalization factor for cell–cell transmission effectiveness.</Annotation>
</Constant>
<Constant symbol="cc_ct_ratio" value="0.9">
<Annotation>Fraction of transmission due to cell–cell contacts vs. extracellular spread.</Annotation>
</Constant>
<Constant symbol="cc_reduced" value="0.2">
<Annotation>Reduction factor for cell–cell transmission.</Annotation>
</Constant>
<Constant symbol="mcs_duration" value="1">
<Annotation>Monte Carlo sampler duration: one cell rearrangement step per simulation time unit.</Annotation>
</Constant>
</Global>
<CellTypes>
<CellType name="medium" class="medium">
<Property symbol="viralsecretion" value="0"/>
</CellType>
<CellType name="basal" class="biological">
<Annotation>Basal stem cells forming the pseudostratified epithelium base, essential for tissue regeneration.</Annotation>
<VolumeConstraint target="60" strength="10">
<Annotation>Low volume constraint reflects thin basal morphology beneath apical layers.</Annotation>
</VolumeConstraint>
<SurfaceConstraint target=".9" strength="10" mode="aspherity">
<Annotation>Controls cell roundness, modeling tight junctions and limited protrusion.</Annotation>
</SurfaceConstraint>
<ConnectivityConstraint>
<Annotation>Enforces cohesive adhesion among basal cells to maintain layer integrity.</Annotation>
</ConnectivityConstraint>
<Function symbol="p_divB">
<Expression>1-exp(-(alphaB*((Nmax-cell_number)/Nmax)))</Expression>
<Annotation>Division probability decreases as total density approaches carrying capacity.</Annotation>
</Function>
<Function symbol="p_deathB">
<Expression>1-exp(-deltaB)</Expression>
<Annotation>Baseline turnover reflecting homeostatic basal cell apoptosis.</Annotation>
</Function>
<Function symbol="p_diffS">
<Expression>1-exp(-diffS)</Expression>
<Annotation>Probabilistic commitment to secretory lineage based on differentiation signal.</Annotation>
</Function>
<AddCell overwrite="true">
<Count>if(cell_number > 25000, celltype.basal.size*p_divB , 0)</Count>
<Distribution>1</Distribution>
<Triggers>
<Rule symbol-ref="Compartment">
<Expression>0.0</Expression>
</Rule>
</Triggers>
<Annotation>Additional basal seeding under high density to maintain layer.</Annotation>
</AddCell>
<CellDivision division-plane="minor">
<Condition>rand_uni(0,1)<p_divB and (cell_number<= 25000)</Condition>
<Triggers>
<Rule symbol-ref="Compartment">
<Expression>0.0</Expression>
</Rule>
</Triggers>
<Annotation>Division along minor axis to mimic observed mitotic orientation.</Annotation>
</CellDivision>
<ChangeCellType time-step="1.0" name="to_secr" newCellType="secretory">
<Condition>Compartment==5</Condition>
<Triggers>
<Rule symbol-ref="Compartment">
<Expression>0</Expression>
</Rule>
</Triggers>
<Annotation>Full differentiation to secretory cell after completing basal compartment cycle.</Annotation>
</ChangeCellType>
<CellDeath>
<Condition>rand_uni(0,1)<p_deathB</Condition>
<Annotation>Random basal cell loss maintaining dynamic equilibrium.</Annotation>
</CellDeath>
<Property symbol="Compartment" value="rand_int(0,4)">
<Annotation>Differentiation stage tracker with four intermediate phases.</Annotation>
</Property>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_diffS</Condition>
<Rule symbol-ref="Compartment">
<Expression>Compartment + 1</Expression>
<Annotation>Advances compartment stage probabilistically per time step.</Annotation>
</Rule>
</Event>
<Property symbol="viralsecretion" value="0">
<Annotation>Ensures no virus release from uninfected basal cells.</Annotation>
</Property>
<ChangeCellType time-step="0.0" newCellType="basal_infected">
<Condition>rand_uni(0,1)<p_infBas</Condition>
<Annotation>Initial infection event based on extracellular viral load.</Annotation>
</ChangeCellType>
<Function symbol="p_infBas">
<Expression>1-exp(-betaB*internal_load)</Expression>
<Annotation>Probability of infection from accumulated internal viral load.</Annotation>
</Function>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
<Annotation>Sums extracellular viral concentrations into per-cell load.</Annotation>
</Mapper>
<Property symbol="internal_load" value="0.0">
<Annotation>Tracks intracellular viral accumulation over time.</Annotation>
</Property>
<NeighborhoodReporter time-step="1.0">
<Input scaling="cell" value="ccbetaB*((cell.type == celltype.basal_infectious.id)*ccrhoB + (cell.type == celltype.secretory_infectious.id)*ccrhoS + (cell.type == celltype.ciliated_infectious.id)*ccrhoC)"/>
<Output symbol-ref="infected_neighbors" mapping="sum"/>
<Annotation>Calculates neighbor-driven infection pressure for secondary spread.</Annotation>
</NeighborhoodReporter>
<Property symbol="infected_neighbors" value="0.0">
<Annotation>Stores count of infectious neighboring cells per time step.</Annotation>
</Property>
<ChangeCellType newCellType="basal_infected">
<Condition>rand_uni(0,1)<1-exp(-infected_neighbors)</Condition>
<Annotation>Secondary infection transformation by cell–cell contact.</Annotation>
</ChangeCellType>
</CellType>
<CellType name="basal_infected" class="biological">
<Annotation>Basal cells in eclipse phase post-infection, prior to viral release. Please find a detailed annoation in "basal".</Annotation>
<Property symbol="t_Bbecomeinfectious" value="0.0"/>
<Function symbol="p_Bbecomeinfectious">
<Expression>1-exp(-kappaB)</Expression>
</Function>
<ChangeCellType time-step="1.0" newCellType="basal_infectious">
<Condition>Compartment == 5</Condition>
</ChangeCellType>
<VolumeConstraint target="60" strength="10"/>
<SurfaceConstraint target=".9" strength="10" mode="aspherity"/>
<Property symbol="viralsecretion" value="0"/>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_Bbecomeinfectious</Condition>
<Rule symbol-ref="Compartment">
<Expression>Compartment + 1</Expression>
</Rule>
</Event>
<Property symbol="Compartment" value="0.0"/>
<ConnectivityConstraint/>
<Property symbol="internal_load" value="0.0"/>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
</Mapper>
</CellType>
<CellType name="basal_infectious" class="biological">
<Annotation>Basal cells actively releasing virus until demise. Please find a detailed annoation in "basal".</Annotation>
<CellDeath>
<Condition>Compartment_d == 5</Condition>
</CellDeath>
<Function symbol="p_deathBInfectious">
<Expression>1-exp(-db_I)</Expression>
</Function>
<VolumeConstraint target="60" strength="10"/>
<SurfaceConstraint target=".9" strength="10" mode="aspherity"/>
<Property symbol="viralsecretion" value="rhoB/cell.volume"/>
<Equation symbol-ref="viralsecretion">
<Expression>rhoB/cell.volume</Expression>
</Equation>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_deathBInfectious</Condition>
<Rule symbol-ref="Compartment_d">
<Expression>Compartment_d + 1</Expression>
</Rule>
</Event>
<Property symbol="Compartment_d" value="0.0"/>
<ConnectivityConstraint/>
<Property symbol="internal_load" value="0.0"/>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
</Mapper>
</CellType>
<CellType name="secretory" class="biological">
<Annotation>Apical secretory cells producing mucus and engaging in viral interactions. Please find a detailed annoation in "basal".</Annotation>
<VolumeConstraint target="60" strength="100"/>
<SurfaceConstraint target=".9" strength="100" mode="aspherity"/>
<ConnectivityConstraint/>
<Function symbol="p_diffCil">
<Expression>1-exp(-diffCil)</Expression>
</Function>
<Function symbol="p_deathS">
<Expression>1-exp(-deltaS)</Expression>
</Function>
<CellDeath>
<Condition>rand_uni(0,1)< p_deathS
</Condition>
</CellDeath>
<ChangeCellType time-step="1.0" name="to_ cil" newCellType="ciliated">
<Condition>Compartment==5</Condition>
</ChangeCellType>
<Property symbol="Compartment" value="rand_int(0,4)"/>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_diffCil</Condition>
<Rule symbol-ref="Compartment">
<Expression>Compartment + 1</Expression>
</Rule>
</Event>
<Property symbol="viralsecretion" value="0"/>
<ChangeCellType time-step="1.0" newCellType="secretory_infected">
<Condition>rand_uni(0,1)<p_infSec</Condition>
</ChangeCellType>
<Function symbol="p_infSec">
<Expression>1-exp(-betaS*internal_load)</Expression>
</Function>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
</Mapper>
<Property symbol="internal_load" value="0.0"/>
<NeighborhoodReporter time-step="1.0">
<Input scaling="cell" value="ccbetaS*((cell.type == celltype.basal_infectious.id)*ccrhoB + (cell.type == celltype.secretory_infectious.id)*ccrhoS + (cell.type == celltype.ciliated_infectious.id)*ccrhoC)"/>
<Output symbol-ref="infected_neighbors" mapping="sum"/>
</NeighborhoodReporter>
<Property symbol="infected_neighbors" value="0.0"/>
<ChangeCellType time-step="1.0" newCellType="secretory_infected">
<Condition>rand_uni(0,1)<1-exp(-infected_neighbors)</Condition>
</ChangeCellType>
</CellType>
<CellType name="secretory_infected" class="biological">
<Annotation>Secretory cells in eclipse phase post-infection, prior to viral release. Please find a detailed annoation in "basal".</Annotation>
<Property symbol="t_Sbecomeinfectious" value="0.0"/>
<Function symbol="p_Sbecomeinfectious">
<Expression>1-exp(-kappaS)</Expression>
</Function>
<ChangeCellType newCellType="secretory_infectious">
<Condition>Compartment == 5</Condition>
</ChangeCellType>
<VolumeConstraint target="60" strength="100"/>
<SurfaceConstraint target=".9" strength="100" mode="aspherity"/>
<Property symbol="viralsecretion" value="0"/>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_Sbecomeinfectious</Condition>
<Rule symbol-ref="Compartment">
<Expression>Compartment + 1</Expression>
</Rule>
</Event>
<Property symbol="Compartment" value="0.0"/>
<ConnectivityConstraint/>
<Property symbol="internal_load" value="0.0"/>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
</Mapper>
</CellType>
<CellType name="secretory_infectious" class="biological">
<Annotation>Secretory cells actively releasing virus until demise. Please find a detailed annoation in "basal".</Annotation>
<VolumeConstraint target="60" strength="100"/>
<Function symbol="p_deathSInfectious">
<Expression>1 - exp(-ds_I)</Expression>
</Function>
<SurfaceConstraint target=".9" strength="100" mode="aspherity"/>
<CellDeath>
<Condition>Compartment_d == 5</Condition>
</CellDeath>
<Property symbol="viralsecretion" value="rhoS/cell.volume"/>
<Equation symbol-ref="viralsecretion">
<Expression>rhoS/cell.volume </Expression>
</Equation>
<Property symbol="Compartment_d" value="0.0"/>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_deathSInfectious</Condition>
<Rule symbol-ref="Compartment_d">
<Expression>Compartment_d + 1</Expression>
</Rule>
</Event>
<ConnectivityConstraint/>
</CellType>
<CellType name="ciliated" class="biological">
<ConnectivityConstraint/>
<SurfaceConstraint target=".9" strength="100" mode="aspherity"/>
<VolumeConstraint target="60" strength="100"/>
<CellDeath>
<Condition>rand_uni(0,1)< p_deathCil
</Condition>
</CellDeath>
<Function symbol="p_deathCil">
<Expression>1-exp(-deltaCil)</Expression>
</Function>
<Property symbol="viralsecretion" value="0"/>
<ChangeCellType time-step="1.0" newCellType="ciliated_infected">
<Condition>rand_uni(0,1)<p_infCil</Condition>
</ChangeCellType>
<Function symbol="p_infCil">
<Expression>1-exp(-betaC*internal_load)</Expression>
</Function>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
</Mapper>
<Property symbol="internal_load" value="0.0"/>
<NeighborhoodReporter time-step="1.0">
<Input scaling="cell" value="ccbetaC*((cell.type == celltype.basal_infectious.id)*ccrhoB + (cell.type == celltype.secretory_infectious.id)*ccrhoS + (cell.type == celltype.ciliated_infectious.id)*ccrhoC)"/>
<Output symbol-ref="infected_neighbors" mapping="sum"/>
</NeighborhoodReporter>
<Property symbol="infected_neighbors" value="0.0"/>
<ChangeCellType time-step="1.0" newCellType="ciliated_infected">
<Condition>rand_uni(0,1)<1-exp(-infected_neighbors)</Condition>
</ChangeCellType>
</CellType>
<CellType name="ciliated_infected" class="biological">
<Annotation>Ciliated cells in eclipse phase post-infection, prior to viral release. Please find a detailed annoation in "basal".</Annotation>
<VolumeConstraint target="60" strength="100"/>
<SurfaceConstraint target=".9" strength="100" mode="aspherity"/>
<ChangeCellType time-step="1.0" newCellType="ciliated_infectious">
<Condition>Compartment == 5</Condition>
</ChangeCellType>
<Function symbol="p_Cbecomeinfectious">
<Expression>1-exp(-kappaC)</Expression>
</Function>
<Property symbol="t_birth_cil_inf" value="0.0"/>
<Property symbol="viralsecretion" value="0"/>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_Cbecomeinfectious</Condition>
<Rule symbol-ref="Compartment">
<Expression>Compartment + 1</Expression>
</Rule>
</Event>
<Property symbol="Compartment" value="0.0"/>
<ConnectivityConstraint/>
<Property symbol="internal_load" value="0.0"/>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
</Mapper>
</CellType>
<CellType name="ciliated_infectious" class="biological">
<Annotation>Ciliated cells actively releasing virus until demise. Please find a detailed annoation in "basal".</Annotation>
<SurfaceConstraint target=".9" strength="100" mode="aspherity"/>
<VolumeConstraint target="60" strength="100"/>
<CellDeath>
<Condition>Compartment_d == 5</Condition>
</CellDeath>
<Function symbol="p_deathCInfectious">
<Expression>1 - exp(-dc_I)</Expression>
</Function>
<Property symbol="t_birth_cil" value="0.0"/>
<Property symbol="viralsecretion" value="rhoC/cell.volume"/>
<Equation symbol-ref="viralsecretion">
<Expression>rhoC/cell.volume </Expression>
</Equation>
<Event trigger="when-true" time-step="1.0">
<Condition>rand_uni(0,1)<p_deathCInfectious</Condition>
<Rule symbol-ref="Compartment_d">
<Expression>Compartment_d + 1</Expression>
</Rule>
</Event>
<Property symbol="Compartment_d" value="0.0"/>
<ConnectivityConstraint/>
<Property symbol="internal_load" value="0.0"/>
<Mapper time-step="1.0" name="internal viral load">
<Input value="ViralLoad"/>
<Output symbol-ref="internal_load" mapping="sum"/>
</Mapper>
</CellType>
</CellTypes>
<CellPopulations>
<Population type="basal" name="basal" size="0">
<InitCircle mode="random" number-of-cells="0.397*init_cell_number">
<Dimensions center="size.x/2,size.y/2,0" radius="size.x/2"/>
</InitCircle>
</Population>
<Population type="secretory" name="secretory" size="0">
<InitCircle name="secretory" mode="random" number-of-cells="0.134*init_cell_number">
<Dimensions center="size.x/2,size.y/2,0" radius="size.x/2"/>
</InitCircle>
</Population>
<Population type="ciliated" name="ciliated" size="0">
<InitCircle name="ciliated" mode="random" number-of-cells="0.469*init_cell_number" tags="">
<Dimensions center="size.x/2,size.y/2,0" radius="size.x/2"/>
</InitCircle>
</Population>
<Population type="basal_infectious" name="basal infectious" size="0">
<InitCircle name="" mode="random" number-of-cells="0" tags="">
<Dimensions center="size.x/2,size.y/2,0" radius="size.x/2"/>
</InitCircle>
</Population>
<Population type="ciliated_infectious" name="ciliated infectious" size="0">
<InitCircle name="" mode="random" number-of-cells="0" tags="">
<Dimensions center="size.x/2,size.y/2,0" radius="size.x/2"/>
</InitCircle>
</Population>
<Population type="secretory_infectious" name="secretory infectious" size="0">
<InitCircle name="" mode="random" number-of-cells="0" tags="">
<Dimensions center="size.x/2,size.y/2,0" radius="size.x/2"/>
</InitCircle>
</Population>
</CellPopulations>
<CPM>
<Interaction>
<Contact type2="secretory" type1="ciliated" value="-20"/>
<Contact type2="ciliated" type1="ciliated" value="10"/>
<Contact type2="secretory" type1="secretory" value="50"/>
<Contact type2="medium" type1="ciliated" value="-10"/>
<Contact type2="medium" type1="secretory" value="-10"/>
<Contact type2="basal" type1="basal" value="50"/>
<Contact type2="ciliated" type1="basal" value="-30"/>
<Contact type2="secretory" type1="basal" value="-30"/>
</Interaction>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>6</Order>
</Neighborhood>
</ShapeSurface>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1"/>
<MetropolisKinetics temperature="37"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
</MonteCarloSampler>
</CPM>
</MorpheusModel>

Downloads
Files associated with this model: