Simplified Actin Waves in Eukaryotic Cell Motility
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Spatial relaxation oscillator produces spatial dynamics and spiral waves, simplification of a previous model for actin waves
Introduction
Motivated by alternative cell motility modes in eukaryotic cells like Dictyostelium discoideum, we consider a simplified version of an intracellular model for an F-actin negative feedback to an (active/inactive) GTPase system. The equations were analysed by full PDE bifurcation analysis in Hughes et al., 2024.
Description
This model is related to a previous model on actin waves.
Equations: for the active ($u$), inactive ($v$) GTPase, respectively, and F-actin ($F$).
$$\begin{align} \frac{\partial u}{\partial t} &= (b+\gamma u^2)v - (1+sF+u^2)u + D \Delta u \\ \frac{\partial v}{\partial t} &= -(b+\gamma u^2)v + (1+sF+u^2)u + \Delta v \\ \frac{\partial F}{\partial t} &= \epsilon (p_0+p_1 u - F) + D_F \Delta F \end{align}$$
The domain is periodic, of (side) length $L$, in 1D and 2D.
The parameters and initial conditions are given in the following table.
Results
One-dimensional analysis
In 1D spatial coordinates we obtain either a polar distribution or a traveling wave, as shown for the variable $u$ in the two kymographs below. Time evolves (horizontal axis) and space is on the vertical axis.
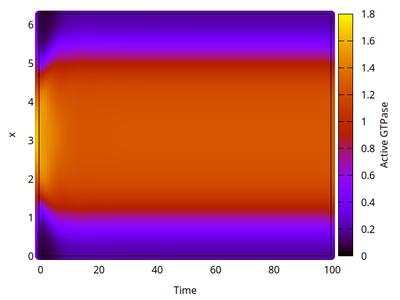
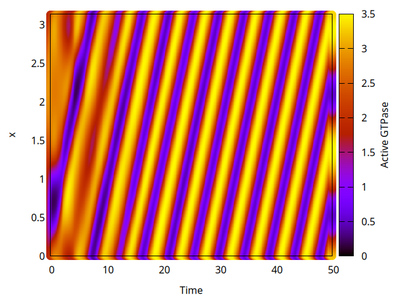
Two-dimensional analysis
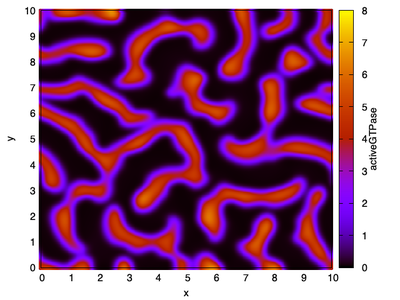
In 2D spatial coordinates, the system can also form dynamic spiral waves as shown in the snapshot and movie below.
Reference
This model is the original used in the publication, up to technical updates:
J. Algorta, A. Fele-Paranj, J. M. Hughes, L. Edelstein-Keshet: Modeling and Simulating Single and Collective Cell Motility. Cold Spring Harb. Perspect. Biol., 2025.
Model
ActinWavesPDE1DPolar_main.xml
XML Preview
<MorpheusModel version="4">
<Description>
<Title>M2071 Actin WavesHughesPolar1D</Title>
<Details>Model ID: https://identifiers.org/morpheus/M2071
File type: Main model
Software: Morpheus (open source). Download from: https://morpheus.gitlab.io
Full title: Simplified Actin Waves in Eukaryotic Cell Motility – Polar 1D
Authors: J. M. Hughes and L. Edelstein-Keshet
Submitter: L. Edelstein-Keshet
Curator: D. Jahn
Date: 29.10.2024
Reference: J. Algorta, A. Fele-Paranj, J. M. Hughes, L. Edelstein-Keshet: Modeling and Simulating Single and Collective Cell Motility. Cold Spring Harb. Perspect. Biol., 2025. https://doi.org/10.1101/cshperspect.a041796
This model reproduces the model and results obtained with Morpheus in Figure 1 of the original publication.
Comment: PDE model for F-actin negative feedback to GTPases, and GTPase promotion of F-actin.
u = active GTPases
v = inactive GTPase
F = F-actin</Details>
</Description>
<!--
Define the variables and initialize their values.
Indicate the rates of diffusion
GTPases diffusem but F-actin does not
-->
<Global>
<Field symbol="u" name="Active GTPase" value="1-0.5* cos(x)">
<Diffusion rate="0.1/dx/dx"/>
</Field>
<Field symbol="v" name="Inactive GTPase" value="1-0.1* cos(x)">
<Diffusion rate="1/dx/dx"/>
</Field>
<Field symbol="F" name="F-actin" value="4.5 + 0.82* cos(x)">
<Diffusion rate="0.001/dx/dx"/>
</Field>
<!--
Specify the method of solution and give the kinetic terms
in the differential equations for the three variables
-->
<System time-step="0.05" solver="Dormand-Prince [adaptive, O(5)]">
<DiffEqn symbol-ref="u">
<Expression> (b+gamma*u^n)*v- u*(1+s*F+u^2) </Expression>
</DiffEqn>
<DiffEqn symbol-ref="v">
<Expression> -(b+gamma*u^n)*v+ u*(1+s*F+u^2) </Expression>
</DiffEqn>
<DiffEqn symbol-ref="F">
<Expression> epsilon*(p0+p1*u-F)</Expression>
</DiffEqn>
<!--
Define the constants and give their values.
-->
<Constant symbol="b" name="basal activation rate" value="0.067"/>
<Constant symbol="gamma" name="autocatalytic activation rate" value="3.55"/>
<Constant symbol="n" name="Hill coefficient" value="2"/>
<Constant symbol="p1" name="GTPase dependent F-actin assembly rate" value="3.8"/>
<Constant symbol="p0" name="F-actin basal growth rate" value="0.8"/>
<Constant symbol="s" name="actin-dependent GTPase inactivation rate" value="0.41"/>
<Constant symbol="epsilon" name="actin reaction rate" value="0.6"/>
</System>
<Function symbol="x">
<Expression>dx*space.x</Expression>
</Function>
<Constant symbol="dx" value="0.02"/>
</Global>
<!--
Specify the 2D Domain ("Square" lattice) and its boundary conditions
Indicate the size of each spatial box
-->
<Space>
<Lattice class="linear">
<Size symbol="size" value="314, 0, 0"/>
<BoundaryConditions>
<Condition type="periodic" boundary="x"/>
<!-- <Disabled>
<Condition type="periodic" boundary="y"/>
</Disabled>
-->
</BoundaryConditions>
<Neighborhood>
<Order>1</Order>
</Neighborhood>
</Lattice>
<SpaceSymbol symbol="space"/>
</Space>
<!--
Set the start and end time and any random seed needed
-->
<Time>
<StartTime value="0"/>
<StopTime value="100"/>
<SaveInterval value="0"/>
<TimeSymbol symbol="time"/>
</Time>
<!--
Specify how the results should be plotted
-->
<Analysis>
<Logger time-step="0.1">
<Input>
<Symbol symbol-ref="u"/>
<Symbol symbol-ref="v"/>
<Symbol symbol-ref="F"/>
<Symbol symbol-ref="x"/>
</Input>
<Output>
<TextOutput file-format="csv"/>
</Output>
<Plots>
<Plot time-step="20">
<Style point-size="2.0" style="points"/>
<Terminal terminal="png"/>
<X-axis minimum="0.0" maximum="100">
<Symbol symbol-ref="time"/>
</X-axis>
<Y-axis minimum="0.0" maximum="6.28">
<Symbol symbol-ref="x"/>
</Y-axis>
<Color-bar>
<Symbol symbol-ref="u"/>
</Color-bar>
</Plot>
</Plots>
</Logger>
<ModelGraph format="dot" reduced="false" include-tags="#untagged"/>
</Analysis>
</MorpheusModel>

Downloads
Files associated with this model: